Chromogenic in situ hybridization is a reliable alternative to fluorescence in situ hybridization for diagnostic testing of 1p and 19q loss in paraffin-embedded gliomas
- PMID: 23107103
- PMCID: PMC8029485
- DOI: 10.1111/bpa.12003
Chromogenic in situ hybridization is a reliable alternative to fluorescence in situ hybridization for diagnostic testing of 1p and 19q loss in paraffin-embedded gliomas
Abstract
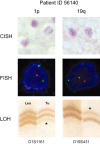
Recent studies imply the importance of rapid and reliable diagnostic assessment of 1p/19q status in oligodendroglial tumors. To date, fluorescence in situ hybridization (FISH) is the most commonly applied technique. FISH, however, has several technical shortcomings that are suboptimal for diagnostic applications: results must be viewed in a fluorescence microscope, results are usually evaluated by a single investigator only, and signal fading excludes physical archiving. Also, in gliomas, the distinction of diffusely infiltrating tumor cells from reactively altered normal tissue may be challenging in fluorescence microscopy. Dual-color chromogenic in situ hybridization (CISH) has started to replace FISH in some diagnostic tests performed in pathology. Here, we present the first single institute experience with a side-by-side analysis of 1p/19q FISH and CISH in a series of 42 consecutive gliomas. FISH and CISH produced identical results for 1p and 19q in 93% of cases (n = 39/42). Discrepant results were reevaluated by repeated FISH and a polymerase chain reaction (PCR)-based microsatellite marker analysis for loss of heterozygosity. Reevaluation confirmed CISH data in all three cases. We conclude that CISH is a reliable alternative in 1p/19q testing in paraffin-embedded tissues likely to be more sensitive to detect 1p/19q status than FISH analysis.
© 2012 The Authors; Brain Pathology © 2012 International Society of Neuropathology.
Figures


Similar articles
-
Is the 1p/19q deletion a diagnostic marker of oligodendrogliomas?Cancer Genet Cytogenet. 2009 Oct;194(1):12-22. doi: 10.1016/j.cancergencyto.2009.05.004. Cancer Genet Cytogenet. 2009. PMID: 19737649
-
Application of brush cytology for FISH-based detection of 1p/19q codeletion in oligodendroglial tumors.J Neurooncol. 2016 Sep;129(3):415-422. doi: 10.1007/s11060-016-2211-0. Epub 2016 Jul 12. J Neurooncol. 2016. PMID: 27406587
-
Next-Generation Sequencing Panel for 1p/19q Codeletion and IDH1-IDH2 Mutational Analysis Uncovers Mistaken Overdiagnoses of 1p/19q Codeletion by FISH.J Mol Diagn. 2021 Sep;23(9):1185-1194. doi: 10.1016/j.jmoldx.2021.06.004. Epub 2021 Jun 26. J Mol Diagn. 2021. PMID: 34186176
-
Molecular diagnostics: techniques and recommendations for 1p/19q assessment.CNS Oncol. 2015;4(5):295-306. doi: 10.2217/cns.15.28. Epub 2015 Nov 6. CNS Oncol. 2015. PMID: 26545171 Free PMC article. Review.
-
The T2-FLAIR-mismatch sign as an imaging biomarker for IDH and 1p/19q status in diffuse low-grade gliomas: a systematic review with a Bayesian approach to evaluation of diagnostic test performance.Neurosurg Focus. 2019 Dec 1;47(6):E13. doi: 10.3171/2019.9.FOCUS19660. Neurosurg Focus. 2019. PMID: 31786548
Cited by
-
Diagnostic accuracy of 1p/19q codeletion tests in oligodendroglioma: A comprehensive meta-analysis based on a Cochrane systematic review.Neuropathol Appl Neurobiol. 2022 Jun;48(4):e12790. doi: 10.1111/nan.12790. Epub 2022 Mar 3. Neuropathol Appl Neurobiol. 2022. PMID: 34958131 Free PMC article.
-
Voxel-wise radiogenomic mapping of tumor location with key molecular alterations in patients with glioma.Neuro Oncol. 2018 Oct 9;20(11):1517-1524. doi: 10.1093/neuonc/noy134. Neuro Oncol. 2018. PMID: 30107597 Free PMC article.
-
Exploiting nanopore sequencing for characterization and grading of IDH-mutant gliomas.Brain Pathol. 2024 Jan;34(1):e13203. doi: 10.1111/bpa.13203. Epub 2023 Aug 13. Brain Pathol. 2024. PMID: 37574201 Free PMC article.
-
Genetic Factors Affecting Intraoperative 5-aminolevulinic Acid-induced Fluorescence of Diffuse Gliomas.Radiol Oncol. 2017 Apr 12;51(2):142-150. doi: 10.1515/raon-2017-0019. eCollection 2017 Jun. Radiol Oncol. 2017. PMID: 28740449 Free PMC article.
-
Multiparametric and multiregional diffusion features help predict molecule information, grade and survival in lower-grade gliomas: a feasibility study.Br J Radiol. 2019 Nov;92(1103):20190324. doi: 10.1259/bjr.20190324. Epub 2019 Aug 9. Br J Radiol. 2019. PMID: 31386559 Free PMC article.
References
-
- Ambros PF, Ambros IM (2001) Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol 37:492–504. - PubMed
-
- Arnould L, Roger P, Macgrogan G, Chenard MP, Balaton A, Beauclair S, Penault‐Llorca F (2012) Accuracy of HER2 status determination on breast core‐needle biopsies (immunohistochemistry, FISH, CISH and SISH vs FISH). Mod Pathol 25:675–682. - PubMed
-
- Asif M, Khadim MT, Mushtaq S, Mamoon N, Akhtar F, Ali Z (2011) Determination of her‐2/neu by chromogenic in situ hybridization on borderline (2+) immunohistochemistry cases in carcinoma breast. Asian Pac J Cancer Prev 12:211–214. - PubMed
-
- Bartlett JM, Campbell FM, Ibrahim M, O'Grady A, Kay E, Faulkes C et al (2011) A UK NEQAS ISH multicenter ring study using the Ventana HER2 dual‐color ISH assay. Am J Clin Pathol 135:157–162. - PubMed
-
- Bernet L, Martinez Benaclocha M, Castera C, Cano Munoz R, Sevilla F, Alba J et al (2012) mRNA in situ hybridization (HistoSonda): a new diagnostic tool for HER2‐status in breast cancer—A multicentric Spanish study. Diagn Mol Pathol 21:84–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

