The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment
- PMID: 23108394
- PMCID: PMC3874579
- DOI: 10.1038/onc.2012.491
The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment
Abstract
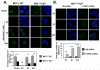
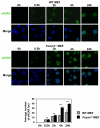
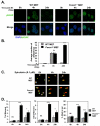
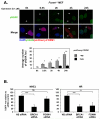
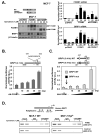
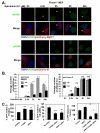
FOXM1 is implicated in genotoxic drug resistance but its role and mechanism of action remain unclear. Here, we establish that γH2AX foci, indicative of DNA double-strand breaks (DSBs), accumulate in a time-dependent manner in the drug-sensitive MCF-7 cells but not in the resistant counterparts in response to epirubicin. We find that FOXM1 expression is associated with epirubicin sensitivity and DSB repair. Ectopic expression of FOXM1 can increase cell viability and abrogate DSBs sustained by MCF-7 cells following epirubicin, owing to an enhancement in repair efficiency. Conversely, alkaline comet and γH2AX foci formation assays show that Foxm1-null cells are hypersensitive to DNA damage, epirubicin and γ-irradiation. Furthermore, we find that FOXM1 is required for DNA repair by homologous recombination (HR) but not non-homologous end joining (NHEJ), using HeLa cell lines harbouring an integrated direct repeat green fluorescent protein reporter for DSB repair. We also identify BRIP1 as a direct transcription target of FOXM1 by promoter analysis and chromatin-immunoprecipitation assay. In agreement, depletion of FOXM1 expression by small interfering RNA downregulates BRIP1 expression at the protein and mRNA levels in MCF-7 and the epirubicin-resistant MCF-7 Epi(R) cells. Remarkably, the requirement for FOXM1 for DSB repair can be circumvented by reintroduction of BRIP1, suggesting that BRIP1 is an important target of FOXM1 in DSB repair. Indeed, like FOXM1, BRIP1 is needed for HR. These data suggest that FOXM1 regulates BRIP1 expression to modulate epirubicin-induced DNA damage repair and drug resistance.
Figures


References
-
- Lin SX, Chen J, Mazumdar M, Poirier D, Wang C, Azzi A, et al. Molecular therapy of breast cancer: progress and future directions. Nat Rev Endocrinol. 2010 Sep;6(9):485–93. - PubMed
-
- Alvarez RH, Valero V, Hortobagyi GN. Emerging targeted therapies for breast cancer. J Clin Oncol. [Research Support, Non-U.S. Gov’t Review] 2010 Jul 10;28(20):3366–79. - PubMed
-
- Zelnak A. Overcoming taxane and anthracycline resistance. Breast J. 2010 May-Jun;16(3):309–12. - PubMed
-
- Stearns V, Davidson NE, Flockhart DA. Pharmacogenetics in the treatment of breast cancer. Pharmacogenomics J. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. Review] 2004;4(3):143–53. - PubMed
-
- Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. [Research Support, Non-U.S. Gov’t Review] 2004 Jun;56(2):185–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

