Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function
- PMID: 23108669
- PMCID: PMC3585520
- DOI: 10.1242/jcs.112631
Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function
Abstract
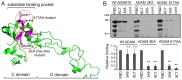
The ADAM10 transmembrane metalloprotease cleaves a variety of cell surface proteins that are important in disease, including ligands for receptor tyrosine kinases of the erbB and Eph families. ADAM10-mediated cleavage of ephrins, the ligands for Eph receptors, is suggested to control Eph/ephrin-mediated cell-cell adhesion and segregation, important during normal developmental processes, and implicated in tumour neo-angiogenesis and metastasis. We previously identified a substrate-binding pocket in the ADAM10 C domain that binds the EphA/ephrin-A complex thereby regulating ephrin cleavage. We have now generated monoclonal antibodies specifically recognising this region of ADAM10, which inhibit ephrin cleavage and Eph/ephrin-mediated cell function, including ephrin-induced Eph receptor internalisation, phosphorylation and Eph-mediated cell segregation. Our studies confirm the important role of ADAM10 in cell-cell interactions mediated by both A- and B-type Eph receptors, and suggest antibodies against the ADAM10 substrate-recognition pocket as promising therapeutic agents, acting by inhibiting cleavage of ephrins and potentially other ADAM10 substrates.
Figures

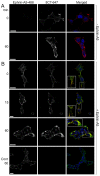
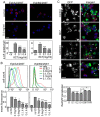
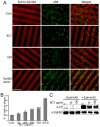
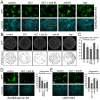
 ) indicates blockade of ephrin cleavage at cell-cell junctions. Ephrin internalisation was calculated using colocalisation of green (ephrin) with red (Eph/293); values are means ± s.e.m., n = 5. *P<0.05, ***P<0.001: significant difference from the untreated or indicated sample. Scale bars: 20 µm.
) indicates blockade of ephrin cleavage at cell-cell junctions. Ephrin internalisation was calculated using colocalisation of green (ephrin) with red (Eph/293); values are means ± s.e.m., n = 5. *P<0.05, ***P<0.001: significant difference from the untreated or indicated sample. Scale bars: 20 µm.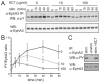
Similar articles
-
Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.Cell. 2005 Oct 21;123(2):291-304. doi: 10.1016/j.cell.2005.08.014. Cell. 2005. PMID: 16239146
-
The role of proteases in regulating Eph/ephrin signaling.Cell Adh Migr. 2014;8(4):294-307. doi: 10.4161/19336918.2014.970026. Cell Adh Migr. 2014. PMID: 25482632 Free PMC article. Review.
-
Therapeutic potential of targeting the Eph/ephrin signaling complex.Int J Biochem Cell Biol. 2018 Dec;105:123-133. doi: 10.1016/j.biocel.2018.10.006. Epub 2018 Oct 19. Int J Biochem Cell Biol. 2018. PMID: 30343150 Free PMC article. Review.
-
Cytoplasmic relaxation of active Eph controls ephrin shedding by ADAM10.PLoS Biol. 2009 Oct;7(10):e1000215. doi: 10.1371/journal.pbio.1000215. Epub 2009 Oct 13. PLoS Biol. 2009. PMID: 19823572 Free PMC article.
-
Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling.Nat Cell Biol. 2011 Jul 31;13(9):1100-7. doi: 10.1038/ncb2298. Nat Cell Biol. 2011. PMID: 21804545
Cited by
-
Spatial organization of EphA2 at the cell-cell interface modulates trans-endocytosis of ephrinA1.Biophys J. 2014 May 20;106(10):2196-205. doi: 10.1016/j.bpj.2014.03.043. Biophys J. 2014. PMID: 24853748 Free PMC article.
-
Role of ADAM10 as a CD30 Sheddase in Classical Hodgkin Lymphoma.Front Immunol. 2020 Mar 31;11:398. doi: 10.3389/fimmu.2020.00398. eCollection 2020. Front Immunol. 2020. PMID: 32296414 Free PMC article. Review.
-
Structural Basis for Regulated Proteolysis by the α-Secretase ADAM10.Cell. 2017 Dec 14;171(7):1638-1648.e7. doi: 10.1016/j.cell.2017.11.014. Epub 2017 Dec 7. Cell. 2017. PMID: 29224781 Free PMC article.
-
Monoclonal antibodies against metzincin targets.Br J Pharmacol. 2019 Jan;176(1):52-66. doi: 10.1111/bph.14186. Epub 2018 Apr 2. Br J Pharmacol. 2019. PMID: 29488211 Free PMC article. Review.
-
ADAM Proteases in Cancer: Biological Roles, Therapeutic Challenges, and Emerging Opportunities.Cancers (Basel). 2025 May 19;17(10):1703. doi: 10.3390/cancers17101703. Cancers (Basel). 2025. PMID: 40427200 Free PMC article. Review.
References
-
- Guan H-H., Goh K-S., Davamani F., Wu P-L., Huang Y-W., Jeyakanthan J., Wu W-G., Chen C-J. (2010). Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins. J. Struct. Biol. 169, 294–303 10.1016/j.jsb.2009.11.009 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

