Differentiating Schistosoma haematobium from related animal schistosomes by PCR amplifying inter-repeat sequences flanking newly selected repeated sequences
- PMID: 23109375
- PMCID: PMC3516075
- DOI: 10.4269/ajtmh.2012.12-0243
Differentiating Schistosoma haematobium from related animal schistosomes by PCR amplifying inter-repeat sequences flanking newly selected repeated sequences
Abstract
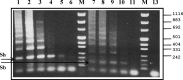
In schistosomiasis elimination programs, successful discrimination of Schistosoma haematobium from the related animal Schistosoma parasites will be essential for accurate detection of human parasite transmission. Polymerase chain reaction assays employing primers from two newly selected repeated sequences, named Sh73 and Sh77, did not discriminate S. haematobium when amplifying Sh73-77 intra- or inter-repeats. However, amplification between Sh73 and the previously described DraI repeat exhibited discriminative banding patterns for S. haematobium and Schistosoma bovis (sensitivity 1 pg and 10 pg, respectively). It also enabled banding pattern discrimination of Schistosoma curassoni and Schistosoma intercalatum, but Schistosoma mattheei and Schistosoma margrebowiei did not yield amplicons. Similar inter-repeat amplification between Sh77 and DraI yielded amplicons with discriminative banding for S. haematobium, and S. bovis; however, S. mattheei was detected only at low sensitivity (1 ng). The Sh73/DraI assay detected snails infected with S. haematobium, S. bovis, or both, and should prove useful for screening snails where discrimination of S. haematobium from related schistosomes is required.
Figures




References
-
- WHO Prevention and control of schistosomiasis and soil transmitted helminthiases: report of a WHO expert committee. World Health Organ Tech Rep Ser. 2002;912:2–3. - PubMed
-
- Hamburger J, He N, Abbasi I, Ramzy RM, Jourdane J, Ruppel A. Polymerase chain reaction assay based on highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome infested water. Am J Trop Med Hyg. 2001;65:907–911. - PubMed
-
- Hamburger J, Hoffman O, Kariuki HC, Muchiri EM, Ouma JH, Koech DK, Sturrock RF, King CH. Large-scale, polymerase chain reaction-based surveillance of Schistosoma haematobium DNA in snails from transmission sites in coastal Kenya: a new tool for studying the dynamics of snail infection. Am J Trop Med Hyg. 2004;71:765–773. - PubMed
-
- Christensen NO, Mutani A, Frandsen F. A review of the biology and transmission ecology of African bovine species of the genus Schistosoma. Z Parasitenkd. 1983;69:551–570. - PubMed
-
- Mone H, Mowahid G, Monard S. The distribution of Schistosoma bovis Sonsino, 1876 in relation to intermediate host mollusc-parasite relationship. Adv Parasitol. 2000;44:100–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

