The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism
- PMID: 23109430
- PMCID: PMC3554121
- DOI: 10.1128/MCB.00881-12
The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism
Abstract
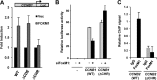
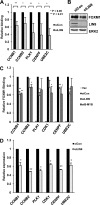
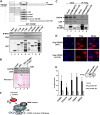
There are nearly 50 forkhead (FOX) transcription factors encoded in the human genome and, due to sharing a common DNA binding domain, they are all thought to bind to similar DNA sequences. It is therefore unclear how these transcription factors are targeted to specific chromatin regions to elicit specific biological effects. Here, we used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to investigate the genome-wide chromatin binding mechanisms used by the forkhead transcription factor FOXM1. In keeping with its previous association with cell cycle control, we demonstrate that FOXM1 binds and regulates a group of genes which are mainly involved in controlling late cell cycle events in the G(2) and M phases. However, rather than being recruited through canonical RYAAAYA forkhead binding motifs, FOXM1 binding is directed via CHR (cell cycle genes homology region) elements. FOXM1 binds these elements through protein-protein interactions with the MMB transcriptional activator complex. Thus, we have uncovered a novel and unexpected mode of chromatin binding of a FOX transcription factor that allows it to specifically control cell cycle-dependent gene expression.
Figures



References
-
- Carlsson P, Mahlapuu M. 2002. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250: 1–23 - PubMed
-
- Breeden LL. 2000. Cyclin transcription: timing is everything. Curr. Biol. 10: R586–R588 - PubMed
-
- Alvarez B, Martinez AC, Burgering BM, Carrera AC. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413: 744–747 - PubMed
-
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7: 126–136 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
