Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus
- PMID: 23109710
- PMCID: PMC3501363
- DOI: 10.1084/jem.20121505
Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus
Abstract
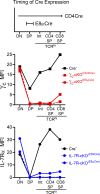
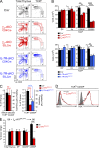
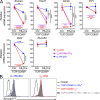
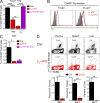
The thymus generates T cells with diverse specificities and functions. To assess the contribution of cytokine receptors to the differentiation of T cell subsets in the thymus, we constructed conditional knockout mice in which IL-7Rα or common cytokine receptor γ chain (γ(c)) genes were deleted in thymocytes just before positive selection. We found that γ(c) expression was required to signal the differentiation of MHC class I (MHC-I)-specific thymocytes into CD8(+) cytotoxic lineage T cells and into invariant natural killer T cells but did not signal the differentiation of MHC class II (MHC-II)-specific thymocytes into CD4(+) T cells, even into regulatory Foxp3(+)CD4(+) T cells which require γ(c) signals for survival. Importantly, IL-7 and IL-15 were identified as the cytokines responsible for CD8(+) cytotoxic T cell lineage specification in vivo. Additionally, we found that small numbers of aberrant CD8(+) T cells expressing Runx3d could arise without γ(c) signaling, but these cells were developmentally arrested before expressing cytotoxic lineage genes. Thus, γ(c)-transduced cytokine signals are required for cytotoxic lineage specification in the thymus and for inducing the differentiation of MHC-I-selected thymocytes into functionally mature T cells.
Figures





Similar articles
-
Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes.J Immunol. 2005 Oct 1;175(7):4465-74. doi: 10.4049/jimmunol.175.7.4465. J Immunol. 2005. PMID: 16177089
-
Identification of lineage-specifying cytokines that signal all CD8+-cytotoxic-lineage-fate 'decisions' in the thymus.Nat Immunol. 2017 Nov;18(11):1218-1227. doi: 10.1038/ni.3847. Epub 2017 Sep 25. Nat Immunol. 2017. PMID: 28945245 Free PMC article.
-
CCR7 expression in developing thymocytes is linked to the CD4 versus CD8 lineage decision.J Immunol. 2007 Dec 1;179(11):7358-64. doi: 10.4049/jimmunol.179.11.7358. J Immunol. 2007. PMID: 18025179
-
CD4/CD8 lineage commitment in T cell receptor transgenic mice: evidence for precommitment of CD4+ CD8+ thymocytes.Semin Immunol. 1994 Aug;6(4):249-56. doi: 10.1006/smim.1994.1032. Semin Immunol. 1994. PMID: 8000034 Review.
-
Adjusting to self in the thymus: CD4 versus CD8 lineage commitment and regulatory T cell development.J Exp Med. 2024 Oct 7;221(10):e20230896. doi: 10.1084/jem.20230896. Epub 2024 Jul 9. J Exp Med. 2024. PMID: 38980291 Free PMC article. Review.
Cited by
-
A Beginner's Guide to T Cell Development.Methods Mol Biol. 2023;2580:3-24. doi: 10.1007/978-1-0716-2740-2_1. Methods Mol Biol. 2023. PMID: 36374448 Review.
-
The Cytokine Receptor IL-7Rα Impairs IL-2 Receptor Signaling and Constrains the In Vitro Differentiation of Foxp3+ Treg Cells.iScience. 2020 Aug 21;23(8):101421. doi: 10.1016/j.isci.2020.101421. Epub 2020 Jul 29. iScience. 2020. PMID: 32791329 Free PMC article.
-
IL-7 induces type 2 cytokine response in lung ILC2s and regulates GATA3 and CD25 expression.J Leukoc Biol. 2022 Nov;112(5):1105-1113. doi: 10.1002/JLB.3AB1220-819RRR. Epub 2022 May 23. J Leukoc Biol. 2022. PMID: 35603486 Free PMC article.
-
From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells.Immunol Res. 2014 Aug;59(1-3):45-55. doi: 10.1007/s12026-014-8545-9. Immunol Res. 2014. PMID: 24847765 Review.
-
PI3K induces B-cell development and regulates B cell identity.Sci Rep. 2018 Jan 22;8(1):1327. doi: 10.1038/s41598-018-19460-5. Sci Rep. 2018. PMID: 29358580 Free PMC article.
References
-
- Bosselut R., Guinter T.I., Sharrow S.O., Singer A. 2003. Unraveling a revealing paradox: Why major histocompatibility complex I-signaled thymocytes “paradoxically” appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J. Exp. Med. 197:1709–1719 10.1084/jem.20030170 - DOI - PMC - PubMed
-
- Brugnera E., Bhandoola A., Cibotti R., Yu Q., Guinter T.I., Yamashita Y., Sharrow S.O., Singer A. 2000. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 13:59–71 10.1016/S1074-7613(00)00008-X - DOI - PubMed
-
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. 2007. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178:280–290 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

