Tat-dependent translocation of an F420-binding protein of Mycobacterium tuberculosis
- PMID: 23110042
- PMCID: PMC3478262
- DOI: 10.1371/journal.pone.0045003
Tat-dependent translocation of an F420-binding protein of Mycobacterium tuberculosis
Abstract
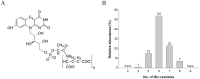
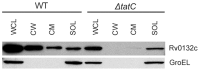
F(420) is a unique cofactor present in a restricted range of microorganisms, including mycobacteria. It has been proposed that F(420) has an important role in the oxidoreductive reactions of Mycobacterium tuberculosis, possibly associated with anaerobic survival and persistence. The protein encoded by Rv0132c has a predicted N-terminal signal sequence and is annotated as an F(420)-dependent glucose-6-phosphate dehydrogenase. Here we show that Rv0132c protein does not have the annotated activity. It does, however, co-purify with F(420) during expression experiments in M. smegmatis. We also show that the Rv0132c-F(420) complex is a substrate for the Tat pathway, which mediates translocation of the complex across the cytoplasmic membrane, where Rv0132c is anchored to the cell envelope. This is the first report of any F(420)-binding protein being a substrate for the Tat pathway and of the presence of F(420) outside of the cytosol in any F(420)-producing microorganism. The Rv0132c protein and its Tat export sequence are essentially invariant in the Mycobacterium tuberculosis complex. Taken together, these results show that current understanding of F(420) biology in mycobacteria should be expanded to include activities occurring in the extra-cytoplasmic cell envelope.
Conflict of interest statement
Figures




References
-
- Global Tuberculosis Control (2010) World Health Organization
-
- Duncan K, Barry CE III (2004) Prospects for new antitubercular drugs. Curr Opin Microbiol 7: 460–465. - PubMed
-
- Bashiri G, Squire CJ, Moreland NJ, Baker EN (2008) Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J Biol Chem 283: 17531–17541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

