Silencing of miR-370 in human cholangiocarcinoma by allelic loss and interleukin-6 induced maternal to paternal epigenotype switch
- PMID: 23110045
- PMCID: PMC3478287
- DOI: 10.1371/journal.pone.0045606
Silencing of miR-370 in human cholangiocarcinoma by allelic loss and interleukin-6 induced maternal to paternal epigenotype switch
Abstract
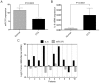
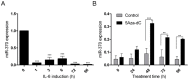
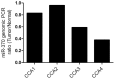
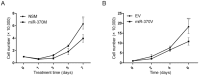
Cholangiocarcinoma (CCA) is a highly lethal malignant tumor arising from the biliary tract epithelium. Interleukin-6 (IL-6) is a major mediator of inflammation and contributor to carcinogenesis within the biliary tree. Previous studies suggested that enforced IL-6 contributes to cholangiocarcinogenesis through hypermethylation of several genes implicated in CCA. However, the precise mechanisms of IL-6 effects in CCA remain unclear. We now demonstrate that microRNA (miR)-370 is underexpressed in a large cohort of human CCA vs. normal liver tissues. In addition, we show that IL-6 induces a time-dependent silencing of miR-370. In addition, demethylation of CCA cells results in upregulation of miR-370. Furthermore, we demonstrate that miR-370 is imprinted, and that the Intergenic Differentially Methylated Region (IG-DMR) responsible for imprinting regulation of this genomic locus is hypermethylated in response to IL-6 treatment. In addition, the IG-DMR is hypermethylated in human CCA specimens compared to normal matched controls, in the same location as the IL-6 induced hypermethylation. Finally, miR-370 was found to regulate WNT10B in luciferase as well as western blotting experiments. Our data indicate that the paternal allele of miR-370 is normally silenced through genomic imprinting and that the overexpression of IL-6 in CCA effectively suppresses the expression of miR-370 from the maternal allele, lending support to the theory that miR-370 silencing in human CCA follows a classic two-hit mechanism.
Conflict of interest statement
Figures



References
-
- Zografos GN, Farfaras A, Zagouri F, Chrysikos D, Karaliotas K (2011) Cholangiocarcinoma: principles and current trends. Hepatobiliary Pancreat Dis Int 10: 10–20. - PubMed
-
- Boumber Y, Issa JP (2011) Epigenetics in cancer: what's the future? Oncology (Williston Park) 25: 220–226, 228. - PubMed
-
- Isomoto H (2009) Epigenetic alterations associated with cholangiocarcinoma (review). Oncol Rep 22: 227–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

