Exclusion of exon 2 is a common mRNA splice variant of primate telomerase reverse transcriptases
- PMID: 23110161
- PMCID: PMC3480478
- DOI: 10.1371/journal.pone.0048016
Exclusion of exon 2 is a common mRNA splice variant of primate telomerase reverse transcriptases
Abstract
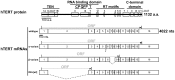
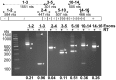
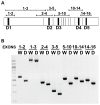
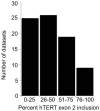
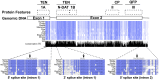
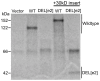
Telomeric sequences are added by an enzyme called telomerase that is made of two components: a catalytic protein called telomerase reverse transcriptase (TERT) and an integral RNA template (TR). Telomerase expression is tightly regulated at each step of gene expression, including alternative splicing of TERT mRNA. While over a dozen different alternative splicing events have been reported for human TERT mRNA, these were all in the 3' half of the coding region. We were interested in examining splicing of the 5' half of hTERT mRNA, especially since exon 2 is unusually large (1.3 kb). Internal mammalian exons are usually short, typically only 50 to 300 nucleotides, and most long internal exons are alternatively processed. We used quantitative RT-PCR and high-throughput sequencing data to examine the variety and quantity of mRNA species generated from the hTERT locus. We determined that there are approximately 20-40 molecules of hTERT mRNA per cell in the A431 human cell line. In addition, we describe an abundant, alternatively-spliced mRNA variant that excludes TERT exon 2 and was seen in other primates. This variant causes a frameshift and results in translation termination in exon 3, generating a 12 kDa polypeptide.
Conflict of interest statement
Figures


Similar articles
-
Regulation of telomerase alternative splicing: a target for chemotherapy.Cell Rep. 2013 Apr 25;3(4):1028-35. doi: 10.1016/j.celrep.2013.03.011. Epub 2013 Apr 4. Cell Rep. 2013. PMID: 23562158 Free PMC article.
-
Human normal T lymphocytes and lymphoid cell lines do express alternative splicing variants of human telomerase reverse transcriptase (hTERT) mRNA.Biochem Biophys Res Commun. 2007 Feb 23;353(4):999-1003. doi: 10.1016/j.bbrc.2006.12.149. Epub 2006 Dec 27. Biochem Biophys Res Commun. 2007. PMID: 17204238
-
Effects of combined siRNA-TR and -TERT on telomerase activity and growth of bladder transitional cell cancer BIU-87 cells.J Huazhong Univ Sci Technolog Med Sci. 2010 Jun;30(3):391-6. doi: 10.1007/s11596-010-0363-2. Epub 2010 Jun 17. J Huazhong Univ Sci Technolog Med Sci. 2010. PMID: 20556588
-
Physiological and pathological significance of human telomerase reverse transcriptase splice variants.Biochimie. 2013 Nov;95(11):1965-70. doi: 10.1016/j.biochi.2013.07.031. Epub 2013 Aug 9. Biochimie. 2013. PMID: 23933091 Review.
-
Alternative Splicing of Human Telomerase Reverse Transcriptase (hTERT) and Its Implications in Physiological and Pathological Processes.Biomedicines. 2021 May 9;9(5):526. doi: 10.3390/biomedicines9050526. Biomedicines. 2021. PMID: 34065134 Free PMC article. Review.
Cited by
-
Insights into Telomerase/hTERT Alternative Splicing Regulation Using Bioinformatics and Network Analysis in Cancer.Cancers (Basel). 2019 May 14;11(5):666. doi: 10.3390/cancers11050666. Cancers (Basel). 2019. PMID: 31091669 Free PMC article. Review.
-
Intronic Cis-Element DR8 in hTERT Is Bound by Splicing Factor SF3B4 and Regulates hTERT Splicing in Non-Small Cell Lung Cancer.Mol Cancer Res. 2022 Oct 4;20(10):1574-1588. doi: 10.1158/1541-7786.MCR-21-0058. Mol Cancer Res. 2022. PMID: 35852380 Free PMC article.
-
Protein kinase C delta (PKCδ) splice variant modulates senescence via hTERT in adipose-derived stem cells.Stem Cell Investig. 2014 Jan 19;1:3. doi: 10.3978/j.issn.2306-9759.2014.01.02. eCollection 2014. Stem Cell Investig. 2014. PMID: 27358850 Free PMC article.
-
Single-cell imaging reveals unexpected heterogeneity of telomerase reverse transcriptase expression across human cancer cell lines.Proc Natl Acad Sci U S A. 2019 Sep 10;116(37):18488-18497. doi: 10.1073/pnas.1908275116. Epub 2019 Aug 26. Proc Natl Acad Sci U S A. 2019. PMID: 31451652 Free PMC article.
-
Avian Leukosis Virus Activation of an Antisense RNA Upstream of TERT in B-Cell Lymphomas.J Virol. 2016 Sep 29;90(20):9509-17. doi: 10.1128/JVI.01127-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27512065 Free PMC article.
References
-
- Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during aging of human fibroblasts. Nature 345(6274): 458–460. - PubMed
-
- Stewart SA, Weinberg RA (2000) Telomerase and human tumorigenesis. Semin Cancer Biol 10(6): 399–406. - PubMed
-
- Morin GB (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59(3): 521–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

