Discovery of bla(OXA-199), a chromosome-based bla(OXA-48)-like variant, in Shewanella xiamenensis
- PMID: 23110226
- PMCID: PMC3480492
- DOI: 10.1371/journal.pone.0048280
Discovery of bla(OXA-199), a chromosome-based bla(OXA-48)-like variant, in Shewanella xiamenensis
Abstract
Introduction: bla(OXA-48) is a globally emerging carbapenemase-encoding gene. The progenitor of bla(OXA-48) appears to be a Shewanella species. The presence of the bla(OXA-48)-like gene was investigated for two Shewanella xiamenensis strains.
Methods: Strain WCJ25 was recovered from post-surgical abdominal drainages, while S4 was the type strain of S. xiamenensis. Species identification for WCJ25 was established by sequencing the 16S rDNA and gyrB genes. PCR was used to screen the bla(OXA-48)-like genes and to obtain their complete sequences. A phylogenetic tree of the bla(OXA-48)-like genes was constructed. The genetic context of the bla(OXA-48)-like gene in strain WCJ25 was investigated by inverse PCR using self-ligated AseI- or RsaI-restricted WCJ25 DNA fragments as template, while that in strain S4 was determined by PCR mapping using that in WCJ25 as template.
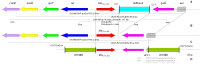
Results: A new bla(OXA-48) variant, designated bla(OXA-48b), with four silent nucleotide differences from the bla(OXA-48) (designated bla(OXA-48a)) found in the Enterobacteriaceae was identified in strain S4. Strain WCJ25 had a new bla(OXA-48)-like variant, bla(OXA-199), with five nucleotide differences from bla(OXA-48)a and bla(OXA-48b). The OXA-199 protein has three amino acid substitutions (H37Y, V44A and D153G) compared with OXA-48. Both bla(OXA-48b) and bla(OXA-199) were found adjacent to genes encoding a peptidase (indicated as orf), a protein of unknown function (sprT), an endonuclease I (endA), and a ribosomal RNA methyl transferase (rsmE) upstream and to transcriptional regulator gene lysR and an acetyl-CoA carboxylase-encoding gene downstream. In addition, the insertion sequence ISShes2 was found inserted downstream of bla(OXA-199) but not of bla(OXA-48b). The 26 bp sequences upstream and 63 bp downstream of bla(OXA-48a), bla(OXA-48b) and bla(OXA-199) were identical.
Conclusions: bla(OXA-48a), bla(OXA-48b) and bla(OXA-199) might have a common origin, suggesting that the bla(OXA-48a) gene found in the Enterobacteriaceae could have originated from the chromosome of S. xiamenensis.
Conflict of interest statement
Figures


References
-
- Poirel L, Potron A, Nordmann P (2012) OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67: 1597–1606. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

