Autoinducers act as biological timers in Vibrio harveyi
- PMID: 23110227
- PMCID: PMC3482212
- DOI: 10.1371/journal.pone.0048310
Autoinducers act as biological timers in Vibrio harveyi
Abstract
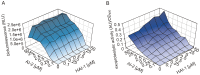
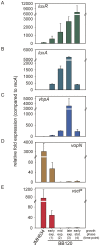
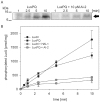
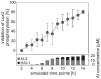
Quorum sensing regulates cell density-dependent phenotypes and involves the synthesis, excretion and detection of so-called autoinducers. Vibrio harveyi strain ATCC BAA-1116 (recently reclassified as Vibrio campbellii), one of the best-characterized model organisms for the study of quorum sensing, produces and responds to three autoinducers. HAI-1, AI-2 and CAI-1 are recognized by different receptors, but all information is channeled into the same signaling cascade, which controls a specific set of genes. Here we examine temporal variations of availability and concentration of the three autoinducers in V. harveyi, and monitor the phenotypes they regulate, from the early exponential to the stationary growth phase in liquid culture. Specifically, the exponential growth phase is characterized by an increase in AI-2 and the induction of bioluminescence, while HAI-1 and CAI-1 are undetectable prior to the late exponential growth phase. CAI-1 activity reaches its maximum upon entry into stationary phase, while molar concentrations of AI-2 and HAI-1 become approximately equal. Similarly, autoinducer-dependent exoproteolytic activity increases at the transition into stationary phase. These findings are reflected in temporal alterations in expression of the luxR gene that encodes the master regulator LuxR, and of four autoinducer-regulated genes during growth. Moreover, in vitro phosphorylation assays reveal a tight correlation between the HAI-1/AI-2 ratio as input and levels of receptor-mediated phosphorylation of LuxU as output. Our study supports a model in which the combinations of autoinducers available, rather than cell density per se, determine the timing of various processes in V. harveyi populations.
Conflict of interest statement
Figures




References
-
- Cano-Gomez A, Høj L, Owens L, Andreakis N (2011) Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst Appl Microbiol 34: 561–565. - PubMed
-
- Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346. - PubMed
-
- Chen X, Schauder S, Potier N, Van DA, Pelczer I, et al. (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415: 545–549. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

