Next generation sequencing in cardiovascular diseases
- PMID: 23110245
- PMCID: PMC3482622
- DOI: 10.4330/wjc.v4.i10.288
Next generation sequencing in cardiovascular diseases
Abstract
In the last few years, the advent of next generation sequencing (NGS) has revolutionized the approach to genetic studies, making whole-genome sequencing a possible way of obtaining global genomic information. NGS has very recently been shown to be successful in identifying novel causative mutations of rare or common Mendelian disorders. At the present time, it is expected that NGS will be increasingly important in the study of inherited and complex cardiovascular diseases (CVDs). However, the NGS approach to the genetics of CVDs represents a territory which has not been widely investigated. The identification of rare and frequent genetic variants can be very important in clinical practice to detect pathogenic mutations or to establish a profile of risk for the development of pathology. The purpose of this paper is to discuss the recent application of NGS in the study of several CVDs such as inherited cardiomyopathies, channelopathies, coronary artery disease and aortic aneurysm. We also discuss the future utility and challenges related to NGS in studying the genetic basis of CVDs in order to improve diagnosis, prevention, and treatment.
Keywords: Cardiomyopathies; Complex disease; Coronary artery disease; Genetics of cardiovascular diseases; Next generation sequencing.
Figures
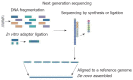
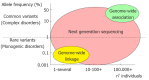
References
-
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. - PubMed
-
- Su Z, Ning B, Fang H, Hong H, Perkins R, Tong W, Shi L. Next-generation sequencing and its applications in molecular diagnostics. Expert Rev Mol Diagn. 2011;11:333–343. - PubMed
-
- Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6–S12. - PubMed
LinkOut - more resources
Full Text Sources

