Overexpression of an isopentenyl diphosphate isomerase gene to enhance trans-polyisoprene production in Eucommia ulmoides Oliver
- PMID: 23110380
- PMCID: PMC3547716
- DOI: 10.1186/1472-6750-12-78
Overexpression of an isopentenyl diphosphate isomerase gene to enhance trans-polyisoprene production in Eucommia ulmoides Oliver
Abstract
Background: Natural rubber produced by plants, known as polyisoprene, is the most widely used isoprenoid polymer. Plant polyisoprenes can be classified into two types; cis-polyisoprene and trans-polyisoprene, depending on the type of polymerization of the isoprene unit. More than 2000 species of higher plants produce latex consisting of cis-polyisoprene. Hevea brasiliensis (rubber tree) produces cis-polyisoprene, and is the key source of commercial rubber. In contrast, relatively few plant species produce trans-polyisoprene. Currently, trans-polyisoprene is mainly produced synthetically, and no plant species is used for its commercial production.
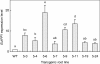
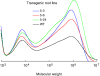
Results: To develop a plant-based system suitable for large-scale production of trans-polyisoprene, we selected a trans-polyisoprene-producing plant, Eucommia ulmoides Oliver, as the target for genetic transformation. A full-length cDNA (designated as EuIPI, Accession No. AB041629) encoding isopentenyl diphosphate isomerase (IPI) was isolated from E. ulmoides. EuIPI consisted of 1028 bp with a 675-bp open reading frame encoding a protein with 224 amino acid residues. EuIPI shared high identity with other plant IPIs, and the recombinant protein expressed in Escherichia coli showed IPI enzymatic activity in vitro. EuIPI was introduced into E. ulmoides via Agrobacterium-mediated transformation. Transgenic lines of E. ulmoides overexpressing EuIPI showed increased EuIPI expression (up to 19-fold that of the wild-type) and a 3- to 4-fold increase in the total content of trans-polyisoprenes, compared with the wild-type (non-transgenic root line) control.
Conclusions: Increasing the expression level of EuIPI by overexpression increased accumulation of trans-polyisoprenes in transgenic E. ulmoides. IPI catalyzes the conversion of isopentenyl diphosphate to its highly electrophilic isomer, dimethylallyl diphosphate, which is the first step in the biosynthesis of all isoprenoids, including polyisoprene. Our results demonstrated that regulation of IPI expression is a key target for efficient production of trans-polyisoprene in E. ulmoides.
Figures





References
-
- Asawatreratanakul K, Zhang YW, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Rattanapittayaporn A, Koyama T. Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis. Eur J Biochem. 2003;270:4671–4680. doi: 10.1046/j.1432-1033.2003.03863.x. - DOI - PubMed
-
- Nakazawa Y, Bamba T, Takeda T, Uefuji H, Harada Y, Li XH, Chen R, Inoue S, Tutumi M, Shimizu T, Su YQ, Gyokusen K, Fukusaki E, Kobayashi A. Production of Eucommia-rubber from Eucommia ulmoides Oliv. (hardy rubber tree) Plant Biot. 2009;26:71–79. doi: 10.5511/plantbiotechnology.26.71. - DOI
-
- Backhaus RA. Rubber formation in plants. Israel J Bot. 1985;34:283–293.
-
- Archer BL, Audley BG. Rubber gutta percha and chicle. Phytochem. 1973;2:310–343.
-
- Schlesinger W, Leeper HM. Chicle - cis- and trans-polyisoprenes from a single plant species. Ind Eng Chem. 1951;43:398–403. doi: 10.1021/ie50494a034. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

