Characterization of a transcriptome from a non-model organism, Cladonia rangiferina, the grey reindeer lichen, using high-throughput next generation sequencing and EST sequence data
- PMID: 23110403
- PMCID: PMC3534622
- DOI: 10.1186/1471-2164-13-575
Characterization of a transcriptome from a non-model organism, Cladonia rangiferina, the grey reindeer lichen, using high-throughput next generation sequencing and EST sequence data
Abstract
Background: Lichens are symbiotic organisms that have a remarkable ability to survive in some of the most extreme terrestrial climates on earth. Lichens can endure frequent desiccation and wetting cycles and are able to survive in a dehydrated molecular dormant state for decades at a time. Genetic resources have been established in lichen species for the study of molecular systematics and their taxonomic classification. No lichen species have been characterised yet using genomics and the molecular mechanisms underlying the lichen symbiosis and the fundamentals of desiccation tolerance remain undescribed. We report the characterisation of a transcriptome of the grey reindeer lichen, Cladonia rangiferina, using high-throughput next-generation transcriptome sequencing and traditional Sanger EST sequencing data.
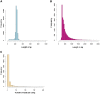
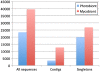
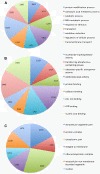
Results: Altogether 243,729 high quality sequence reads were de novo assembled into 16,204 contigs and 49,587 singletons. The genome of origin for the sequences produced was predicted using Eclat with sequences derived from the axenically grown symbiotic partners used as training sequences for the classification model. 62.8% of the sequences were classified as being of fungal origin while the remaining 37.2% were predicted as being of algal origin. The assembled sequences were annotated by BLASTX comparison against a non-redundant protein sequence database with 34.4% of the sequences having a BLAST match. 29.3% of the sequences had a Gene Ontology term match and 27.9% of the sequences had a domain or structural match following an InterPro search. 60 KEGG pathways with more than 10 associated sequences were identified.
Conclusions: Our results present a first transcriptome sequencing and de novo assembly for a lichen species and describe the ongoing molecular processes and the most active pathways in C. rangiferina. This brings a meaningful contribution to publicly available lichen sequence information. These data provide a first glimpse into the molecular nature of the lichen symbiosis and characterise the transcriptional space of this remarkable organism. These data will also enable further studies aimed at deciphering the genetic mechanisms behind lichen desiccation tolerance.
Figures

References
-
- Ahmadjian V. The lichen symbiosis. New York: John Wiley & Sons, Inc.; 1993.
-
- Honegger R. Functional-Aspects of the Lichen Symbiosis. Annual Review of Plant Physiology and Plant Molecular Biology. 1991;42:553–578. doi: 10.1146/annurev.pp.42.060191.003005. - DOI
-
- Collins CR, Farrar JF. Structural resistance to mass transfer in the lichen Xanthoria parietina. New Phytol. 1978;81:71–83. doi: 10.1111/j.1469-8137.1978.tb01605.x. - DOI
-
- Hawksworth DL, Kirk PM, Sutton BC, Pegler DN. Dictionary of the fungi. Wallingford: CAB; 1995.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

