Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps
- PMID: 23110894
- PMCID: PMC3517249
- DOI: 10.1105/tpc.112.103002
Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps
Abstract
Chloroplast RNA metabolism is mediated by a multitude of nuclear encoded factors, many of which are highly specific for individual RNA processing events. In addition, a family of chloroplast ribonucleoproteins (cpRNPs) has been suspected to regulate larger sets of chloroplast transcripts. This together with their propensity for posttranslational modifications in response to external cues suggested a potential role of cpRNPs in the signal-dependent coregulation of chloroplast genes. We show here on a transcriptome-wide scale that the Arabidopsis thaliana cpRNPs CP31A and CP29A (for 31 kD and 29 kD chloroplast protein, respectively), associate with large, overlapping sets of chloroplast transcripts. We demonstrate that both proteins are essential for resistance of chloroplast development to cold stress. They are required to guarantee transcript stability of numerous mRNAs at low temperatures and under these conditions also support specific processing steps. Fine mapping of cpRNP-RNA interactions in vivo suggests multiple points of contact between these proteins and their RNA ligands. For CP31A, we demonstrate an essential function in stabilizing sense and antisense transcripts that span the border of the small single copy region and the inverted repeat of the chloroplast genome. CP31A associates with the common 3'-terminus of these RNAs and protects them against 3'-exonucleolytic activity.
Figures
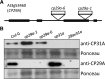

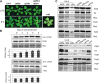




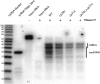
References
-
- Churin Y., Hess W.R., Börner T. (1999). Cloning and characterization of three cDNAs encoding chloroplast RNA-binding proteins from barley (Hordeum vulgare L.): Differential regulation of expression by light and plastid development. Curr. Genet. 36: 173–181 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

