PNA-based microRNA inhibitors elicit anti-inflammatory effects in microglia cells
- PMID: 23111503
- PMCID: PMC3593954
- DOI: 10.1039/c2cc36540e
PNA-based microRNA inhibitors elicit anti-inflammatory effects in microglia cells
Abstract
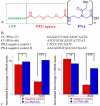
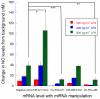
Peptide nucleic acid (PNA) inhibitors of miR-221-3p (CU-PNA-221) and miR-466l-3p (CU-PNA-466) demonstrated changes in inflammatory responses. Suppression of inflammatory signalling was unexpected and further investigation led to the identification of calmodulin as a novel target of miRNA-466l-3p. These studies demonstrate that exogenous agents may suppress neuroinflammation mediated by microglial cells.
Figures
Similar articles
-
Benfotiamine attenuates inflammatory response in LPS stimulated BV-2 microglia.PLoS One. 2015 Feb 19;10(2):e0118372. doi: 10.1371/journal.pone.0118372. eCollection 2015. PLoS One. 2015. PMID: 25695433 Free PMC article.
-
Anti-Inflammatory and Anti-Migratory Activities of Isoquinoline-1-Carboxamide Derivatives in LPS-Treated BV2 Microglial Cells via Inhibition of MAPKs/NF-κB Pathway.Int J Mol Sci. 2020 Mar 27;21(7):2319. doi: 10.3390/ijms21072319. Int J Mol Sci. 2020. PMID: 32230861 Free PMC article.
-
Modelling neuroinflammation in vitro: a tool to test the potential neuroprotective effect of anti-inflammatory agents.PLoS One. 2012;7(9):e45227. doi: 10.1371/journal.pone.0045227. Epub 2012 Sep 20. PLoS One. 2012. PMID: 23028862 Free PMC article.
-
Anti-inflammatory mechanisms of N-adamantyl-4-methylthiazol-2-amine in lipopolysaccharide-stimulated BV-2 microglial cells.Int Immunopharmacol. 2014 Sep;22(1):73-83. doi: 10.1016/j.intimp.2014.06.022. Epub 2014 Jun 25. Int Immunopharmacol. 2014. PMID: 24975832
-
Inhibition of neuroinflammation by thymoquinone requires activation of Nrf2/ARE signalling.Int Immunopharmacol. 2017 Jul;48:17-29. doi: 10.1016/j.intimp.2017.04.018. Epub 2017 May 3. Int Immunopharmacol. 2017. PMID: 28458100
Cited by
-
Inhibition of MicroRNA-221 Alleviates Neuropathic Pain Through Targeting Suppressor of Cytokine Signaling 1.J Mol Neurosci. 2016 Jul;59(3):411-20. doi: 10.1007/s12031-016-0748-1. Epub 2016 Apr 8. J Mol Neurosci. 2016. PMID: 27059231
-
Balancing the immune response in the brain: IL-10 and its regulation.J Neuroinflammation. 2016 Nov 24;13(1):297. doi: 10.1186/s12974-016-0763-8. J Neuroinflammation. 2016. PMID: 27881137 Free PMC article. Review.
-
Targeting miR‑155‑5p and miR‑221‑3p by peptide nucleic acids induces caspase‑3 activation and apoptosis in temozolomide‑resistant T98G glioma cells.Int J Oncol. 2019 Jul;55(1):59-68. doi: 10.3892/ijo.2019.4810. Epub 2019 May 23. Int J Oncol. 2019. PMID: 31180529 Free PMC article.
-
Synthesis of PNA oligoether conjugates.Molecules. 2014 Mar 13;19(3):3135-48. doi: 10.3390/molecules19033135. Molecules. 2014. PMID: 24633349 Free PMC article.
-
Synergistic effects of the combined treatment of U251 and T98G glioma cells with an anti‑tubulin tetrahydrothieno[2,3‑c]pyridine derivative and a peptide nucleic acid targeting miR‑221‑3p.Int J Oncol. 2021 Aug;59(2):61. doi: 10.3892/ijo.2021.5241. Epub 2021 Jul 19. Int J Oncol. 2021. PMID: 34278445 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

