Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites
- PMID: 23112148
- PMCID: PMC3503224
- DOI: 10.1073/pnas.1201400109
Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites
Abstract
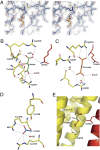
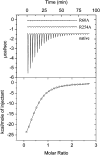
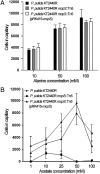
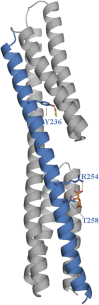
Chemoreceptor-based signaling is a central mechanism in bacterial signal transduction. Receptors are classified according to the size of their ligand-binding region. The well-studied cluster I proteins have a 100- to 150-residue ligand-binding region that contains a single site for chemoattractant recognition. Cluster II receptors, which contain a 220- to 300-residue ligand-binding region and which are almost as abundant as cluster I receptors, remain largely uncharacterized. Here, we report high-resolution structures of the ligand-binding region of the cluster II McpS chemotaxis receptor (McpS-LBR) of Pseudomonas putida KT2440 in complex with different chemoattractants. The structure of McpS-LBR represents a small-molecule binding domain composed of two modules, each able to bind different signal molecules. Malate and succinate were found to bind to the membrane-proximal module, whereas acetate binds to the membrane-distal module. A structural alignment of the two modules revealed that the ligand-binding sites could be superimposed and that amino acids involved in ligand recognition are conserved in both binding sites. Ligand binding to both modules was shown to trigger chemotactic responses. Further analysis showed that McpS-like receptors were found in different classes of proteobacteria, indicating that this mode of response to different carbon sources may be universally distributed. The physiological relevance of the McpS architecture may lie in its capacity to respond with high sensitivity to the preferred carbon sources malate and succinate and, at the same time, mediate lower sensitivity responses to the less preferred but very abundant carbon source acetate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Crystallization and crystallographic analysis of the ligand-binding domain of the Pseudomonas putida chemoreceptor McpS in complex with malate and succinate.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Apr 1;68(Pt 4):428-31. doi: 10.1107/S1744309112004940. Epub 2012 Mar 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22505412 Free PMC article.
-
Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands.J Biol Chem. 2010 Jul 23;285(30):23126-36. doi: 10.1074/jbc.M110.110403. Epub 2010 May 24. J Biol Chem. 2010. PMID: 20498372 Free PMC article.
-
Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions.Environ Microbiol. 2010 Nov;12(11):2873-84. doi: 10.1111/j.1462-2920.2010.02325.x. Epub 2010 Aug 25. Environ Microbiol. 2010. PMID: 20738376 Review.
-
Physiologically relevant divalent cations modulate citrate recognition by the McpS chemoreceptor.J Mol Recognit. 2011 Mar-Apr;24(2):378-85. doi: 10.1002/jmr.1101. J Mol Recognit. 2011. PMID: 21360620
-
Chemoreceptor-based signal sensing.Curr Opin Biotechnol. 2017 Jun;45:8-14. doi: 10.1016/j.copbio.2016.11.021. Epub 2017 Jan 11. Curr Opin Biotechnol. 2017. PMID: 28088095 Review.
Cited by
-
Bacterial sensor evolved by decreasing complexity.Proc Natl Acad Sci U S A. 2025 Feb 4;122(5):e2409881122. doi: 10.1073/pnas.2409881122. Epub 2025 Jan 29. Proc Natl Acad Sci U S A. 2025. PMID: 39879239 Free PMC article.
-
The role of solute binding proteins in signal transduction.Comput Struct Biotechnol J. 2021 Mar 26;19:1786-1805. doi: 10.1016/j.csbj.2021.03.029. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33897981 Free PMC article. Review.
-
Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium.Cell Host Microbe. 2015 Aug 12;18(2):147-56. doi: 10.1016/j.chom.2015.07.002. Cell Host Microbe. 2015. PMID: 26269952 Free PMC article.
-
Desiccation-induced viable but nonculturable state in Pseudomonas putida KT2440, a survival strategy.PLoS One. 2019 Jul 19;14(7):e0219554. doi: 10.1371/journal.pone.0219554. eCollection 2019. PLoS One. 2019. PMID: 31323038 Free PMC article.
-
Pseudomonas aeruginosa as a Model To Study Chemosensory Pathway Signaling.Microbiol Mol Biol Rev. 2021 Jan 13;85(1):e00151-20. doi: 10.1128/MMBR.00151-20. Print 2021 Feb 17. Microbiol Mol Biol Rev. 2021. PMID: 33441490 Free PMC article. Review.
References
-
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5(11):862–872. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

