CDX2 is an amplified lineage-survival oncogene in colorectal cancer
- PMID: 23112155
- PMCID: PMC3503165
- DOI: 10.1073/pnas.1206004109
CDX2 is an amplified lineage-survival oncogene in colorectal cancer
Abstract
The mutational activation of oncogenes drives cancer development and progression. Classic oncogenes, such as MYC and RAS, are active across many different cancer types. In contrast, "lineage-survival" oncogenes represent a distinct and emerging class typically comprising transcriptional regulators of a specific cell lineage that, when deregulated, support the proliferation and survival of cancers derived from that lineage. Here, in a large collection of colorectal cancer cell lines and tumors, we identify recurrent amplification of chromosome 13, an alteration highly restricted to colorectal-derived cancers. A minimal region of amplification on 13q12.2 pinpoints caudal type homeobox transcription factor 2 (CDX2), a regulator of normal intestinal lineage development and differentiation, as a target of the amplification. In contrast to its described role as a colorectal tumor suppressor, CDX2 when amplified is required for the proliferation and survival of colorectal cancer cells. Further, transcriptional profiling, binding-site analysis, and functional studies link CDX2 to Wnt/β-catenin signaling, itself a key oncogenic pathway in colorectal cancer. These data characterize CDX2 as a lineage-survival oncogene deregulated in colorectal cancer. Our findings challenge a prevailing view that CDX2 is a tumor suppressor in colorectal cancer and uncover an additional piece in the multistep model of colorectal tumorigenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



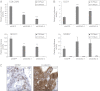
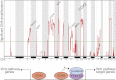
References
-
- American Cancer Society (2011) Cancer Facts & Figures 2011 (American Cancer Society, Atlanta)
-
- Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. - PubMed
-
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. - PubMed
-
- Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: A meta-analysis. Gut. 2008;57(7):941–950. - PubMed
-
- Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41(14):2060–2070. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

