Alkyltransferase-like protein (Atl1) distinguishes alkylated guanines for DNA repair using cation-π interactions
- PMID: 23112169
- PMCID: PMC3503161
- DOI: 10.1073/pnas.1209451109
Alkyltransferase-like protein (Atl1) distinguishes alkylated guanines for DNA repair using cation-π interactions
Abstract
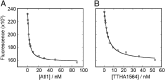
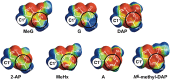
Alkyltransferase-like (ATL) proteins in Schizosaccharomyces pombe (Atl1) and Thermus thermophilus (TTHA1564) protect against the adverse effects of DNA alkylation damage by flagging O(6)-alkylguanine lesions for nucleotide excision repair (NER). We show that both ATL proteins bind with high affinity to oligodeoxyribonucleotides containing O(6)-alkylguanines differing in size, polarity, and charge of the alkyl group. However, Atl1 shows a greater ability than TTHA1564 to distinguish between O(6)-alkylguanine and guanine and in an unprecedented mechanism uses Arg69 to probe the electrostatic potential surface of O(6)-alkylguanine, as determined using molecular mechanics calculations. An unexpected consequence of this feature is the recognition of 2,6-diaminopurine and 2-aminopurine, as confirmed in crystal structures of respective Atl1-DNA complexes. O(6)-Alkylguanine and guanine discrimination is diminished for Atl1 R69A and R69F mutants, and S. pombe R69A and R69F mutants are more sensitive toward alkylating agent toxicity, revealing the key role of Arg69 in identifying O(6)-alkylguanines critical for NER recognition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


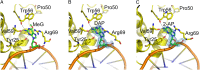

Similar articles
-
Atl1 regulates choice between global genome and transcription-coupled repair of O(6)-alkylguanines.Mol Cell. 2012 Jul 13;47(1):50-60. doi: 10.1016/j.molcel.2012.04.028. Epub 2012 May 31. Mol Cell. 2012. PMID: 22658721 Free PMC article.
-
A novel DNA damage recognition protein in Schizosaccharomyces pombe.Nucleic Acids Res. 2006 May 5;34(8):2347-54. doi: 10.1093/nar/gkl270. Print 2006. Nucleic Acids Res. 2006. PMID: 16679453 Free PMC article.
-
Structural basis of O6-alkylguanine recognition by a bacterial alkyltransferase-like DNA repair protein.J Biol Chem. 2010 Apr 30;285(18):13736-41. doi: 10.1074/jbc.M109.093591. Epub 2010 Mar 8. J Biol Chem. 2010. PMID: 20212037 Free PMC article.
-
Alkyltransferase-like proteins.DNA Repair (Amst). 2007 Aug 1;6(8):1222-8. doi: 10.1016/j.dnarep.2007.03.014. Epub 2007 May 17. DNA Repair (Amst). 2007. PMID: 17500045 Review.
-
Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O(6)-alkylguanine-DNA alkyltransferase.Mutat Res. 2000 Aug 30;460(3-4):151-63. doi: 10.1016/s0921-8777(00)00024-0. Mutat Res. 2000. PMID: 10946226 Review.
Cited by
-
A combination of direct reversion and nucleotide excision repair counters the mutagenic effects of DNA carboxymethylation.DNA Repair (Amst). 2022 Feb;110:103262. doi: 10.1016/j.dnarep.2021.103262. Epub 2021 Dec 29. DNA Repair (Amst). 2022. PMID: 35030424 Free PMC article.
-
Bracken Fern Carcinogen, Ptaquiloside, Forms a Guanine O6-Adduct in DNA.J Agric Food Chem. 2025 Jan 15;73(2):1053-1061. doi: 10.1021/acs.jafc.4c07187. Epub 2025 Jan 7. J Agric Food Chem. 2025. PMID: 39772526 Free PMC article.
-
The Cation-π Interaction in Chemistry and Biology.Chem Rev. 2025 Mar 12;125(5):2793-2808. doi: 10.1021/acs.chemrev.4c00707. Epub 2025 Feb 20. Chem Rev. 2025. PMID: 39977669 Free PMC article. Review.
-
Biochemical and structural studies of the Mycobacterium tuberculosis O6-methylguanine methyltransferase and mutated variants.J Bacteriol. 2013 Jun;195(12):2728-36. doi: 10.1128/JB.02298-12. Epub 2013 Apr 5. J Bacteriol. 2013. PMID: 23564173 Free PMC article.
-
Probing DNA by 2-OG-dependent dioxygenase.Cell. 2013 Dec 19;155(7):1448-50. doi: 10.1016/j.cell.2013.12.002. Cell. 2013. PMID: 24360270 Free PMC article.
References
-
- Margison GP, Santibáñez-Koref MF. O6-alkylguanine-DNA alkyltransferase: Role in carcinogenesis and chemotherapy. Bioessays. 2002;24(3):255–266. - PubMed
-
- Daniels DS, et al. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11(8):714–720. - PubMed
-
- McMurry TBH. MGMT inhibitors—The Trinity College-Paterson Institute experience, a chemist’s perception. DNA Repair (Amst) 2007;6(8):1161–1169. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

