In vivo architecture and action of bacterial structural maintenance of chromosome proteins
- PMID: 23112333
- PMCID: PMC3807729
- DOI: 10.1126/science.1227126
In vivo architecture and action of bacterial structural maintenance of chromosome proteins
Abstract
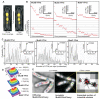
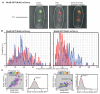
SMC (structural maintenance of chromosome) proteins act ubiquitously in chromosome processing. In Escherichia coli, the SMC complex MukBEF plays roles in chromosome segregation and organization. We used single-molecule millisecond multicolor fluorescence microscopy of live bacteria to reveal that a dimer of dimeric fluorescent MukBEF molecules acts as the minimal functional unit. On average, 8 to 10 of these complexes accumulated as "spots" in one to three discrete chromosome-associated regions of the cell, where they formed higher-order structures. Functional MukBEF within spots exchanged with freely diffusing complexes at a rate of one complex about every 50 seconds in reactions requiring adenosine triphosphate (ATP) hydrolysis. Thus, by functioning in pairs, MukBEF complexes may undergo multiple cycles of ATP hydrolysis without being released from DNA, analogous to the behavior of well-characterized molecular motors.
Figures

References
-
- Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Mol Microbiol. 2011;81:855. - PubMed
-
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595. - PubMed
-
- Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

