Evidence for a Contribution of ALA Synthesis to Plastid-To-Nucleus Signaling
- PMID: 23112801
- PMCID: PMC3483025
- DOI: 10.3389/fpls.2012.00236
Evidence for a Contribution of ALA Synthesis to Plastid-To-Nucleus Signaling
Abstract
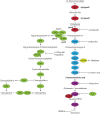
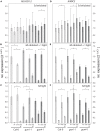
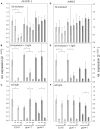
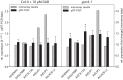
The formation of 5-aminolevulinic acid (ALA) in tetrapyrrole biosynthesis is widely controlled by environmental and metabolic feedback cues that determine the influx into the entire metabolic path. Because of its central role as the rate-limiting step, we hypothesized a potential role of ALA biosynthesis in tetrapyrrole-mediated retrograde signaling and exploited the direct impact of ALA biosynthesis on nuclear gene expression (NGE) by using two different approaches. Firstly, the Arabidopsisgun1, hy1 (gun2), hy2 (gun3), gun4 mutants showing uncoupled NGE from the physiological state of chloroplasts were thoroughly examined for regulatory modifications of ALA synthesis and transcriptional control in the nucleus. We found that reduced ALA-synthesizing capacity is common to analyzed gun mutants. Inhibition of ALA synthesis by gabaculine (GAB) that inactivates glutamate-1-semialdehyde aminotransferase and ALA feeding of wild-type and mutant seedlings corroborate the expression data of gun mutants. Transcript level of photosynthetic marker genes were enhanced in norflurazon (NF)-treated seedlings upon additional GAB treatment, while enhanced ALA amounts diminish these RNA levels in NF-treated wild-type in comparison to the solely NF-treated seedlings. Secondly, the impact of posttranslationally down-regulated ALA synthesis on NGE was investigated by global transcriptome analysis of GAB-treated Arabidopsis seedlings and the gun4-1 mutant, which is also characterized by reduced ALA formation. A common set of significantly modulated genes was identified indicating ALA synthesis as a potential signal emitter. The over-represented gene ontology categories of genes with decreased or increased transcript abundance highlight a few biological processes and cellular functions, which are remarkably affected in response to plastid-localized ALA biosynthesis. These results support the hypothesis that ALA biosynthesis correlates with retrograde signaling-mediated control of NGE.
Keywords: ALA synthesis; gabaculine; gun mutants; microarray analysis; retrograde signaling.
Figures





Similar articles
-
In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling.Physiol Plant. 2010 Apr;138(4):503-19. doi: 10.1111/j.1399-3054.2009.01343.x. Epub 2009 Dec 17. Physiol Plant. 2010. PMID: 20028479
-
Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling.Plant J. 2000 Dec;24(6):883-94. doi: 10.1046/j.1365-313x.2000.00936.x. Plant J. 2000. PMID: 11135121
-
Tetrapyrrole biosynthesis pathway regulates plastid-to-nucleus signaling by controlling plastid gene expression in plants.Plant Commun. 2023 Jan 9;4(1):100411. doi: 10.1016/j.xplc.2022.100411. Epub 2022 Jul 14. Plant Commun. 2023. PMID: 35836377 Free PMC article.
-
The Role of Tetrapyrrole- and GUN1-Dependent Signaling on Chloroplast Biogenesis.Plants (Basel). 2021 Jan 21;10(2):196. doi: 10.3390/plants10020196. Plants (Basel). 2021. PMID: 33494334 Free PMC article. Review.
-
Coordination of plastid and nuclear gene expression.Philos Trans R Soc Lond B Biol Sci. 2003 Jan 29;358(1429):135-44; discussion 144-5. doi: 10.1098/rstb.2002.1180. Philos Trans R Soc Lond B Biol Sci. 2003. PMID: 12594922 Free PMC article. Review.
Cited by
-
A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis.J Integr Plant Biol. 2016 Jan;58(1):91-103. doi: 10.1111/jipb.12369. Epub 2015 Jul 24. J Integr Plant Biol. 2016. PMID: 25989254 Free PMC article.
-
The essential role of sugar metabolism in the acclimation response of Arabidopsis thaliana to high light intensities.J Exp Bot. 2014 Apr;65(6):1619-36. doi: 10.1093/jxb/eru027. Epub 2014 Feb 12. J Exp Bot. 2014. PMID: 24523502 Free PMC article.
-
The xantha Marker Trait Is Associated with Altered Tetrapyrrole Biosynthesis and Deregulated Transcription of PhANGs in Rice.Front Plant Sci. 2017 May 31;8:901. doi: 10.3389/fpls.2017.00901. eCollection 2017. Front Plant Sci. 2017. PMID: 28620402 Free PMC article.
-
Changes in plastid biogenesis leading to the formation of albino regenerants in barley microspore culture.BMC Plant Biol. 2021 Jan 7;21(1):22. doi: 10.1186/s12870-020-02755-z. BMC Plant Biol. 2021. PMID: 33413097 Free PMC article.
-
A Suppressor Mutation Partially Reverts the xantha Trait via Lowered Methylation in the Promoter of Genomes Uncoupled 4 in Rice.Front Plant Sci. 2019 Aug 2;10:1003. doi: 10.3389/fpls.2019.01003. eCollection 2019. Front Plant Sci. 2019. PMID: 31428119 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

