One-step Agrobacterium mediated transformation of eight genes essential for rhizobium symbiotic signaling using the novel binary vector system pHUGE
- PMID: 23112864
- PMCID: PMC3480454
- DOI: 10.1371/journal.pone.0047885
One-step Agrobacterium mediated transformation of eight genes essential for rhizobium symbiotic signaling using the novel binary vector system pHUGE
Abstract
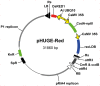
Advancement in plant research is becoming impaired by the fact that the transfer of multiple genes is difficult to achieve. Here we present a new binary vector for Agrobacterium tumefaciens mediated transformation, pHUGE-Red, in concert with a cloning strategy suited for the transfer of up to nine genes at once. This vector enables modular cloning of large DNA fragments by employing Gateway technology and contains DsRED1 as visual selection marker. Furthermore, an R/Rs inducible recombination system was included allowing subsequent removal of the selection markers in the newly generated transgenic plants. We show the successful use of pHUGE-Red by transferring eight genes essential for Medicago truncatula to establish a symbiosis with rhizobia bacteria as one 74 kb T-DNA into four non-leguminous species; strawberry, poplar, tomato and tobacco. We provide evidence that all transgenes are expressed in the root tissue of the non-legumes. Visual control during the transformation process and subsequent marker gene removal makes the pHUGE-Red vector an excellent tool for the efficient transfer of multiple genes.
Conflict of interest statement
Figures





Similar articles
-
Medicago truncatula transformation using root explants.Methods Mol Biol. 2006;343:137-42. doi: 10.1385/1-59745-130-4:137. Methods Mol Biol. 2006. PMID: 16988340 Review.
-
A Unified Agrobacterium-Mediated Transformation Protocol for Alfalfa (Medicago sativa L.) and Medicago truncatula.Methods Mol Biol. 2019;1864:153-163. doi: 10.1007/978-1-4939-8778-8_11. Methods Mol Biol. 2019. PMID: 30415335
-
Transgenic Medicago truncatula plants obtained from Agrobacterium tumefaciens -transformed roots and Agrobacterium rhizogenes-transformed hairy roots.Planta. 2006 May;223(6):1344-54. doi: 10.1007/s00425-006-0268-2. Epub 2006 Mar 31. Planta. 2006. PMID: 16575594
-
Medicago truncatula transformation using cotyledon explants.Methods Mol Biol. 2006;343:129-35. doi: 10.1385/1-59745-130-4:129. Methods Mol Biol. 2006. PMID: 16988339 Review.
-
Backbone-free transformation of barrel medic (Medicago truncatula) with a Medicago-derived transfer DNA.Plant Cell Rep. 2010 Sep;29(9):1013-21. doi: 10.1007/s00299-010-0887-8. Epub 2010 Jun 23. Plant Cell Rep. 2010. PMID: 20571798
Cited by
-
Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses.Proc Natl Acad Sci U S A. 2018 May 15;115(20):E4700-E4709. doi: 10.1073/pnas.1721395115. Epub 2018 May 1. Proc Natl Acad Sci U S A. 2018. PMID: 29717040 Free PMC article.
-
Inactivation of Three Stacked Genes of Cytosolic Peroxiredoxins by Genome Editing.Methods Mol Biol. 2024;2798:235-263. doi: 10.1007/978-1-0716-3826-2_17. Methods Mol Biol. 2024. PMID: 38587748
-
Plant Biosystems Design Research Roadmap 1.0.Biodes Res. 2020 Dec 5;2020:8051764. doi: 10.34133/2020/8051764. eCollection 2020. Biodes Res. 2020. PMID: 37849899 Free PMC article. Review.
-
Traffic lines: new tools for genetic analysis in Arabidopsis thaliana.Genetics. 2015 May;200(1):35-45. doi: 10.1534/genetics.114.173435. Epub 2015 Feb 23. Genetics. 2015. PMID: 25711279 Free PMC article.
-
Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops.BMC Biol. 2019 Dec 3;17(1):99. doi: 10.1186/s12915-019-0710-0. BMC Biol. 2019. PMID: 31796086 Free PMC article. Review.
References
-
- Charpentier M, Oldroyd G (2010) How close are we to nitrogen-fixing cereals? Curr Opin Plant Biol. 13: 556–64. Review. - PubMed
-
- Burrill TJ, Hansen R (1917) Is symbiosis possible between legume bacteria and non-legume plants? Urbana, Ill.: University of Illinois Agricultural Experiment Station, University of Illinois. Agricultural Experiment Station 202: 161–181.
-
- Vainstein A, Marton I, Zuker A, Danziger M, Tzfira T (2011) Permanent genome modifications in plant cells by transient viral vectors. Trends Biotechnol. 29: 363–9. - PubMed
-
- Miao Y, Jiang L (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc. 2: 2348–53. - PubMed
-
- Taylor NJ, Fauquet CM (2002) Microparticle bombardment as a tool in plant science and agricultural biotechnology. DNA Cell Biol. 21: 963–77. Review. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

