Calculating standard reduction potentials of [4Fe-4S] proteins
- PMID: 23115132
- PMCID: PMC3570669
- DOI: 10.1002/jcc.23169
Calculating standard reduction potentials of [4Fe-4S] proteins
Abstract
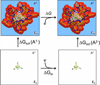
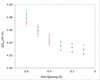
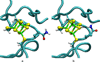
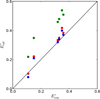
The oxidation-reduction potentials of electron transfer proteins determine the driving forces for their electron transfer reactions. Although the type of redox site determines the intrinsic energy required to add or remove an electron, the electrostatic interaction energy between the redox site and its surrounding environment can greatly shift the redox potentials. Here, a method for calculating the reduction potential versus the standard hydrogen electrode, E°, of a metalloprotein using a combination of density functional theory and continuum electrostatics is presented. This work focuses on the methodology for the continuum electrostatics calculations, including various factors that may affect the accuracy. The calculations are demonstrated using crystal structures of six homologous HiPIPs, which give E° that are in excellent agreement with experimental results.
Copyright © 2012 Wiley Periodicals, Inc.
Figures
Similar articles
-
Fold versus sequence effects on the driving force for protein-mediated electron transfer.Proteins. 2010 Oct;78(13):2798-808. doi: 10.1002/prot.22794. Proteins. 2010. PMID: 20635418 Free PMC article.
-
Insight into environmental effects on bonding and redox properties of [4Fe-4S] clusters in proteins.J Am Chem Soc. 2009 Apr 29;131(16):5724-5. doi: 10.1021/ja900406j. J Am Chem Soc. 2009. PMID: 19341280 Free PMC article.
-
Probing ligand effects on the redox energies of [4Fe-4S] clusters using broken-symmetry density functional theory.J Phys Chem A. 2009 May 14;113(19):5671-6. doi: 10.1021/jp809446q. J Phys Chem A. 2009. PMID: 19378988 Free PMC article.
-
Structure and electrochemistry of proteins harboring iron-sulfur clusters of different nuclearities. Part I. [4Fe-4S]+[2Fe-2S] iron-sulfur proteins.J Struct Biol. 2017 Oct;200(1):1-19. doi: 10.1016/j.jsb.2017.05.010. Epub 2017 Jun 13. J Struct Biol. 2017. PMID: 28619651 Review.
-
Structural principles for computational and de novo design of 4Fe-4S metalloproteins.Biochim Biophys Acta. 2016 May;1857(5):531-538. doi: 10.1016/j.bbabio.2015.10.001. Epub 2015 Oct 9. Biochim Biophys Acta. 2016. PMID: 26449207 Free PMC article. Review.
Cited by
-
Theoretical Modeling of Redox Potentials of Biomolecules.Molecules. 2022 Feb 5;27(3):1077. doi: 10.3390/molecules27031077. Molecules. 2022. PMID: 35164342 Free PMC article. Review.
-
Identifying residues that cause pH-dependent reduction potentials.Biochemistry. 2013 May 7;52(18):3022-4. doi: 10.1021/bi4002858. Epub 2013 Apr 24. Biochemistry. 2013. PMID: 23607577 Free PMC article.
-
Prediction of Reduction Potentials of Copper Proteins with Continuum Electrostatics and Density Functional Theory.Chemistry. 2017 Nov 2;23(61):15436-15445. doi: 10.1002/chem.201702901. Epub 2017 Sep 21. Chemistry. 2017. PMID: 28815759 Free PMC article.
-
Quantum Mechanical Calculations of Redox Potentials of the Metal Clusters in Nitrogenase.Molecules. 2022 Dec 21;28(1):65. doi: 10.3390/molecules28010065. Molecules. 2022. PMID: 36615260 Free PMC article.
-
Benchmark Study of Redox Potential Calculations for Iron-Sulfur Clusters in Proteins.Inorg Chem. 2022 Apr 25;61(16):5991-6007. doi: 10.1021/acs.inorgchem.1c03422. Epub 2022 Apr 11. Inorg Chem. 2022. PMID: 35403427 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources