A critical assessment of feature selection methods for biomarker discovery in clinical proteomics
- PMID: 23115301
- PMCID: PMC3536906
- DOI: 10.1074/mcp.M112.022566
A critical assessment of feature selection methods for biomarker discovery in clinical proteomics
Abstract
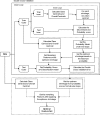
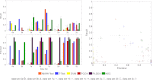
In this paper, we compare the performance of six different feature selection methods for LC-MS-based proteomics and metabolomics biomarker discovery-t test, the Mann-Whitney-Wilcoxon test (mww test), nearest shrunken centroid (NSC), linear support vector machine-recursive features elimination (SVM-RFE), principal component discriminant analysis (PCDA), and partial least squares discriminant analysis (PLSDA)-using human urine and porcine cerebrospinal fluid samples that were spiked with a range of peptides at different concentration levels. The ideal feature selection method should select the complete list of discriminating features that are related to the spiked peptides without selecting unrelated features. Whereas many studies have to rely on classification error to judge the reliability of the selected biomarker candidates, we assessed the accuracy of selection directly from the list of spiked peptides. The feature selection methods were applied to data sets with different sample sizes and extents of sample class separation determined by the concentration level of spiked compounds. For each feature selection method and data set, the performance for selecting a set of features related to spiked compounds was assessed using the harmonic mean of the recall and the precision (f-score) and the geometric mean of the recall and the true negative rate (g-score). We conclude that the univariate t test and the mww test with multiple testing corrections are not applicable to data sets with small sample sizes (n = 6), but their performance improves markedly with increasing sample size up to a point (n > 12) at which they outperform the other methods. PCDA and PLSDA select small feature sets with high precision but miss many true positive features related to the spiked peptides. NSC strikes a reasonable compromise between recall and precision for all data sets independent of spiking level and number of samples. Linear SVM-RFE performs poorly for selecting features related to the spiked compounds, even though the classification error is relatively low.
Figures



References
-
- Mischak H., Allmaier G., Apweiler R., Attwood T., Baumann M., Benigni A., Bennett S. E., Bischoff R., Bongcam-Rudloff E., Capasso G., Coon J. J., D'Haese P., Dominiczak A. F., Dakna M., Dihazi H., Ehrich J. H., Fernandez-Llama P., Fliser D., Frokiaer J., Garin J., Girolami M., Hancock W. S., Haubitz M., Hochstrasser D., Holman R. R., Ioannidis J. P., Jankowski J., Julian B. A., Klein J. B., Kolch W., Luider T., Massy Z., Mattes W. B., Molina F., Monsarrat B., Novak J., Peter K., Rossing P., Sanchez-Carbayo M., Schanstra J. P., Semmes O. J., Spasovski G., Theodorescu D., Thongboonkerd V., Vanholder R., Veenstra T. D., Weissinger E., Yamamoto T., Vlahou A. (2010) Recommendations for biomarker identification and qualification in clinical proteomics. Sci. Transl. Med. 2, 46ps42 - PubMed
-
- Puntmann V. O. (2009) How-to guide on biomarkers: biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad. Med. J. 85, 538–545 - PubMed
-
- Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 - PubMed
-
- Saeys Y., Inza I., Larranaga P. (2007) A review of feature selection techniques in bioinformatics. Bioinformatics 23, 2507–2517 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

