Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies
- PMID: 23115339
- PMCID: PMC3511153
- DOI: 10.1073/pnas.1217207109
Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies
Abstract
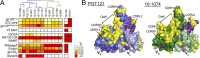
Broadly neutralizing HIV antibodies (bNAbs) can recognize carbohydrate-dependent epitopes on gp120. In contrast to previously characterized glycan-dependent bNAbs that recognize high-mannose N-glycans, PGT121 binds complex-type N-glycans in glycan microarrays. We isolated the B-cell clone encoding PGT121, which segregates into PGT121-like and 10-1074-like groups distinguished by sequence, binding affinity, carbohydrate recognition, and neutralizing activity. Group 10-1074 exhibits remarkable potency and breadth but no detectable binding to protein-free glycans. Crystal structures of unliganded PGT121, 10-1074, and their likely germ-line precursor reveal that differential carbohydrate recognition maps to a cleft between complementarity determining region (CDR)H2 and CDRH3. This cleft was occupied by a complex-type N-glycan in a "liganded" PGT121 structure. Swapping glycan contact residues between PGT121 and 10-1074 confirmed their importance for neutralization. Although PGT121 binds complex-type N-glycans, PGT121 recognized high-mannose-only HIV envelopes in isolation and on virions. As HIV envelopes exhibit varying proportions of high-mannose- and complex-type N-glycans, these results suggest promiscuous carbohydrate interactions, an advantageous adaptation ensuring neutralization of all viruses within a given strain.
Conflict of interest statement
Conflict of interest statement: M.C.N., H.M., P.J.B. and L.S. have a pending patent application for the new PGT121 antibody variants described in the present study with the United States Patent and Trademark Office.
Figures




References
-
- Plotkin SA. Vaccines: Correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47(3):401–409. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

