Computational medicine: translating models to clinical care
- PMID: 23115356
- PMCID: PMC3618897
- DOI: 10.1126/scitranslmed.3003528
Computational medicine: translating models to clinical care
Abstract
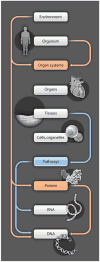
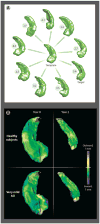
Because of the inherent complexity of coupled nonlinear biological systems, the development of computational models is necessary for achieving a quantitative understanding of their structure and function in health and disease. Statistical learning is applied to high-dimensional biomolecular data to create models that describe relationships between molecules and networks. Multiscale modeling links networks to cells, organs, and organ systems. Computational approaches are used to characterize anatomic shape and its variations in health and disease. In each case, the purposes of modeling are to capture all that we know about disease and to develop improved therapies tailored to the needs of individuals. We discuss advances in computational medicine, with specific examples in the fields of cancer, diabetes, cardiology, and neurology. Advances in translating these computational methods to the clinic are described, as well as challenges in applying models for improving patient health.
Figures


References
-
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. - PubMed
-
- Dolmetsch R. Excitation-transcription coupling: Signaling by ion channels to the nucleus. Sci STKE. 2003;2003:PE4. - PubMed
-
- Frank D, Kuhn C, Brors B, Hanselmann C, Lüdde M, Katus HA, Frey N. Gene expression pattern in biomechanically stretched cardiomyocytes: Evidence for a stretch-specific gene program. Hypertension. 2008;51:309–318. - PubMed
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

