The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas
- PMID: 23116066
- PMCID: PMC3515803
- DOI: 10.1186/1476-4598-11-82
The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas
Abstract
Background: Despite continuous efforts to identify genes that are pivotal regulators of advanced melanoma and closely related to it, to determine which of these genes have to be blocked in their function to keep this highly aggressive disease in check, it is far from clear which molecular pathway(s) and specific genes therein, is the Achilles' heel of primary and metastatic melanoma. In this report, we present data, which document that the DEAD-box helicase DDX11, which is required for sister chromatid cohesion, is a crucial gatekeeper for melanoma cell survival.
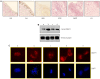
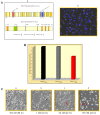
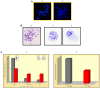
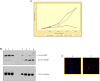
Methods: Performing immunohistochemistry and immunoblot analysis, we determined expression of DDX11 in melanoma tissues and cell lines. Following transfection of melanoma cells with a DDX11-specific siRNA, we conducted a qPCR analysis to determine downregulation of DDX11 in the transfected melanoma cells. In subsequent studies, which focused upon an analysis of fluorescently labeled as well as Giesma-stained chromosome spreads, a proliferation analysis and apoptosis assays, we determined the impact of suppressing DDX11 expression on melanoma cells representing advanced melanoma.
Result: The findings of the study presented herein document that DDX11 is upregulated with progression from noninvasive to invasive melanoma, and that it is expressed at high levels in advanced melanoma. Furthermore, and equally important, we demonstrate that blocking the expression of DDX11 leads not only to inhibition of melanoma cell proliferation and severe defects in chromosome segregation, but also drives melanoma cells rapidly into massive apoptosis.
Conclusion: To date, little is known as to whether helicases play a role in melanoma development and specifically, in the progression from early to advanced melanoma. In this report, we show that the helicase DDX11 is expressed at high levels in primary and metastatic melanoma, and that interfering with its expression leads to severe chromosome segregation defects, telomere shortening, and massive melanoma cell apoptosis. These findings suggest that DDX11 could be an important candidate for molecular targeted therapy for advanced melanoma.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

