The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites
- PMID: 23118352
- PMCID: PMC3542864
- DOI: 10.1093/hmg/dds465
The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites
Abstract
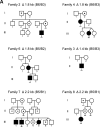
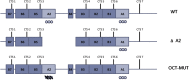
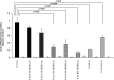
At chromosome 11p15.5, the imprinting centre 1 (IC1) controls the parent of origin-specific expression of the IGF2 and H19 genes. The 5 kb IC1 region contains multiple target sites (CTS) for the zinc-finger protein CTCF, whose binding on the maternal chromosome prevents the activation of IGF2 and allows that of H19 by common enhancers. CTCF binding helps maintaining the maternal IC1 methylation-free, whereas on the paternal chromosome gamete-inherited DNA methylation inhibits CTCF interaction and enhancer-blocking activity resulting in IGF2 activation and H19 silencing. Maternally inherited 1.4-2.2 kb deletions are associated with methylation of the residual CTSs and Beckwith-Wiedemann syndrome, although with different penetrance and expressivity. We explored the relationship between IC1 microdeletions and phenotype by analysing a number of previously described and novel mutant alleles. We used a highly quantitative assay based on next generation sequencing to measure DNA methylation in affected families and analysed enhancer-blocking activity and CTCF binding in cultured cells. We demonstrate that the microdeletions mostly affect IC1 function and CTCF binding by changing CTS spacing. Thus, the extent of IC1 inactivation and the clinical phenotype are influenced by the arrangement of the residual CTSs. A CTS spacing similar to the wild-type allele results in moderate IC1 inactivation and is associated with stochastic DNA methylation of the maternal IC1 and incomplete penetrance. Microdeletions with different CTS spacing display severe IC1 inactivation and are associated with IC1 hypermethylation and complete penetrance. Careful characterization of the IC1 microdeletions is therefore needed to predict recurrence risks and phenotypical outcomes.
Figures



References
-
- Choufani S., Shuman C., Weksberg R. Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2010;154C:343–354. - PubMed
-
- Cerrato F., Sparago A., Di Matteo I., Zou X., Dean W., Sasaki H., Smith P., Genesio R., Bruggemann M., Reik W., Riccio A. The two-domain hypothesis in Beckwith–Wiedemann syndrome: autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Hum. Mol. Genet. 2005a;14:503–511. - PubMed
-
- Ferguson-Smith A.C. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011;12:565–575. - PubMed
-
- Bartolomei M.S., Ferguson-Smith A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011;3 doi:10.1101/cshperspect.a002592. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

