Signal-BNF: a Bayesian network fusing approach to predict signal peptides
- PMID: 23118510
- PMCID: PMC3481251
- DOI: 10.1155/2012/492174
Signal-BNF: a Bayesian network fusing approach to predict signal peptides
Abstract
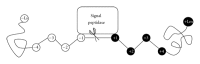
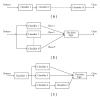
A signal peptide is a short peptide chain that directs the transport of a protein and has become the crucial vehicle in finding new drugs or reprogramming cells for gene therapy. As the avalanche of new protein sequences generated in the postgenomic era, the challenge of identifying new signal sequences has become even more urgent and critical in biomedical engineering. In this paper, we propose a novel predictor called Signal-BNF to predict the N-terminal signal peptide as well as its cleavage site based on Bayesian reasoning network. Signal-BNF is formed by fusing the results of different Bayesian classifiers which used different feature datasets as its input through weighted voting system. Experiment results show that Signal-BNF is superior to the popular online predictors such as Signal-3L and PrediSi. Signal-BNF is featured by high prediction accuracy that may serve as a useful tool for further investigating many unclear details regarding the molecular mechanism of the zip code protein-sorting system in cells.
Figures



Similar articles
-
Signal-3L: A 3-layer approach for predicting signal peptides.Biochem Biophys Res Commun. 2007 Nov 16;363(2):297-303. doi: 10.1016/j.bbrc.2007.08.140. Epub 2007 Aug 31. Biochem Biophys Res Commun. 2007. PMID: 17880924
-
Prediction of protein signal sequences and their cleavage sites by statistical rulers.Biochem Biophys Res Commun. 2005 Dec 16;338(2):1005-11. doi: 10.1016/j.bbrc.2005.10.046. Epub 2005 Oct 21. Biochem Biophys Res Commun. 2005. PMID: 16256954
-
Predicting secretory protein signal sequence cleavage sites by fusing the marks of global alignments.Amino Acids. 2007;32(4):493-6. doi: 10.1007/s00726-006-0466-z. Epub 2006 Nov 15. Amino Acids. 2007. PMID: 17103116
-
Prediction of protein signal sequences.Curr Protein Pept Sci. 2002 Dec;3(6):615-22. doi: 10.2174/1389203023380468. Curr Protein Pept Sci. 2002. PMID: 12470215 Review.
-
Architecture, function and prediction of long signal peptides.Brief Bioinform. 2009 Sep;10(5):569-78. doi: 10.1093/bib/bbp030. Epub 2009 Jun 17. Brief Bioinform. 2009. PMID: 19535397 Review.
Cited by
-
Prediction of Signal Peptides in Proteins from Malaria Parasites.Int J Mol Sci. 2018 Nov 22;19(12):3709. doi: 10.3390/ijms19123709. Int J Mol Sci. 2018. PMID: 30469512 Free PMC article.
-
In silico high throughput mutagenesis and screening of signal peptides to mitigate N-terminal heterogeneity of recombinant monoclonal antibodies.MAbs. 2022 Jan-Dec;14(1):2044977. doi: 10.1080/19420862.2022.2044977. MAbs. 2022. PMID: 35275041 Free PMC article.
-
A survey of computational intelligence techniques in protein function prediction.Int J Proteomics. 2014;2014:845479. doi: 10.1155/2014/845479. Epub 2014 Dec 11. Int J Proteomics. 2014. PMID: 25574395 Free PMC article. Review.
References
-
- Chou K-C. Prediction of protein signal sequences. Current Protein and Peptide Science. 2002;3(6):615–622. - PubMed
-
- Chou K-C. Structural bioinformatics and its impact to biomedical science. Current Medicinal Chemistry. 2004;11(16):2105–2134. - PubMed
-
- Lubec G, Afjehi-Sadat L, Yang JW, John JPP. Searching for hypothetical proteins: theory and practice based upon original data and literature. Progress in Neurobiology. 2005;77(1-2):90–127. - PubMed
-
- Shen H-B, Chou K-C. Signal-3L: a 3-layer approach for predicting signal peptides. Biochemical and Biophysical Research Communications. 2007;363(2):297–303. - PubMed
-
- Chou K-C, Shen H-B. Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochemical and Biophysical Research Communications. 2007;357(3):633–640. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

