Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato
- PMID: 23118951
- PMCID: PMC3485194
- DOI: 10.1371/journal.pone.0048198
Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato
Abstract
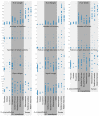
Tomato, Solanum lycopersicum, is divided into two widely distributed varieties: the cultivated S. lycopersicum var. lycopersicum, and the weedy S. lycopersicum var. cerasiforme. Solanum pimpinellifolium is the most closely related wild species of tomato.The roles of S. pimpinellifolium and S. l. cerasiforme during the domestication of tomato are still under debate. Some authors consider S. l. cerasiforme to be the ancestor, whereas others think that S. l. cerasiforme is an admixture of S. pimpinellifolium and the cultivated S. l. lycopersicum. It is also not clear whether the domestication occurred in the Andean region or in Mesoamerica. We characterized 272 accessions (63 S. pimpinellifolium, 106 S. l. cerasiforme, 95 S. l. lycopersicum and 8 derived from hybridization processes) were morphologically and genetically using the SolCap platform (7,414 SNPs). The two species were distinguished in a PCA analysis and displayed a rich geographic structure. Solanum lycopersicum var. cerasiforme and S. l. lycopersicum were also differentiated in the PCA and Structure analyses, which supports maintaining them as different varieties. Solanum pimpinellifolium and the Andean S. l. cerasiforme were more diverse than the non-Andean S. lycopersicum. Solanum lycopersicum var. cerasiforme was morphologically and molecularly intermediate between S. pimpinellifolium and tomato. Solanum lycopersicum var. cerasiforme, with the exception of several Ecuadorian and Mexican accessions, is composed of the products of admixture processes according to the Structure analysis. The non-admixtured S. l. cerasiforme might be similar to the ancestral cultivars from which the cultivated tomato originated, and presents remarkable morphological diversity, including fruits of up to 6 cm in diameter. The data obtained would fit a model in which a pre-domestication took place in the Andean region, with the domestication being completed in Mesoamerica. Subsequently, the Spaniards took plants from Mesoamerica to Spain and from there they were exported to the rest of the world.
Conflict of interest statement
Figures





References
-
- Peralta IE, Spooner DM, Knapp S (2008) Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae). Ann Arbor, Mi.: American Society of Plant Taxonomists. 186 p., 181 lám. col. p.
-
- Warnock SJ (1991) Natural habitats of Lycopersicon species. HortScience 26: 466–471.
-
- Miller JC, Tanksley SD (1990) RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon . Theoretical and applied genetics 80: 437–448. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

