PairMotif: A new pattern-driven algorithm for planted (l, d) DNA motif search
- PMID: 23119020
- PMCID: PMC3485246
- DOI: 10.1371/journal.pone.0048442
PairMotif: A new pattern-driven algorithm for planted (l, d) DNA motif search
Abstract
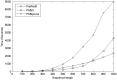
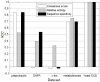
Motif search is a fundamental problem in bioinformatics with an important application in locating transcription factor binding sites (TFBSs) in DNA sequences. The exact algorithms can report all (l, d) motifs and find the best one under a specific objective function. However, it is still a challenging task to identify weak motifs, since either a large amount of memory or execution time is required by current exact algorithms. A new exact algorithm, PairMotif, is proposed for planted (l, d) motif search (PMS) in this paper. To effectively reduce both candidate motifs and scanned l-mers, multiple pairs of l-mers with relatively large distances are selected from input sequences to restrict the search space. Comparisons with several recently proposed algorithms show that PairMotif requires less storage space and runs faster on most PMS instances. Particularly, among the algorithms compared, only PairMotif can solve the weak instance (27, 9) within 10 hours. Moreover, the performance of PairMotif is stable over the sequence length, which allows it to identify motifs in longer sequences. For the real biological data, experimental results demonstrate the validity of the proposed algorithm.
Conflict of interest statement
Figures

References
-
- Pevzner PA, Sze SH (2000) Combinatorial approaches to finding subtle signals in DNA sequences. In: Altman R, Bailey TL, eds. Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology. California: AAAI Press. 269–278. - PubMed
-
- Boucher C, Brown DG, Church P (2007) A graph clustering approach to weak motif recognition. In: Giancarlo R, Hannenhalli S, eds. Proceedings of the 7th International Workshop on Algorithms in Bioinformatics. Philadelphia: LNCS. 149–160.
-
- Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, et al. (1993) Detecting subtle sequence signals: a Gibb’s sampling strategy for multiple alignment. Science 262: 208–214. - PubMed
-
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R, Brutlag D, eds. Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. California: AAAI Press. 28–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

