Scaffold-based pan-agonist design for the PPARα, PPARβ and PPARγ receptors
- PMID: 23119024
- PMCID: PMC3485212
- DOI: 10.1371/journal.pone.0048453
Scaffold-based pan-agonist design for the PPARα, PPARβ and PPARγ receptors
Abstract
As important members of nuclear receptor superfamily, Peroxisome proliferator-activated receptors (PPAR) play essential roles in regulating cellular differentiation, development, metabolism, and tumorigenesis of higher organisms. The PPAR receptors have 3 identified subtypes: PPARα, PPARβ and PPARγ, all of which have been treated as attractive targets for developing drugs to treat type 2 diabetes. Due to the undesirable side-effects, many PPAR agonists including PPARα/γ and PPARβ/γ dual agonists are stopped by US FDA in the clinical trials. An alternative strategy is to design novel pan-agonist that can simultaneously activate PPARα, PPARβ and PPARγ. Under such an idea, in the current study we adopted the core hopping algorithm and glide docking procedure to generate 7 novel compounds based on a typical PPAR pan-agonist LY465608. It was observed by the docking procedures and molecular dynamics simulations that the compounds generated by the core hopping and glide docking not only possessed the similar functions as the original LY465608 compound to activate PPARα, PPARβ and PPARγ receptors, but also had more favorable conformation for binding to the PPAR receptors. The additional absorption, distribution, metabolism and excretion (ADME) predictions showed that the 7 compounds (especially Cpd#1) hold high potential to be novel lead compounds for the PPAR pan-agonist. Our findings can provide a new strategy or useful insights for designing the effective pan-agonists against the type 2 diabetes.
Conflict of interest statement
Figures
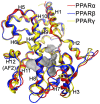

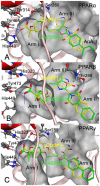
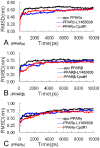
References
-
- Wild S, Roglic G, Green A, Sicree R, King H (2004) Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27: 1047–1053. - PubMed
-
- Zimmet P, Shaw J, Alberti KG (2003) Preventing Type 2 diabetes and the dysmetabolic syndrome in the real world: a realistic view. Diabet Med 20: 693–702. - PubMed
-
- Berger J, Moller DE (2002) The mechanisms of action of PPARs. Annu Rev Med 53: 409–435. - PubMed
-
- Berger JP, Akiyama TE, Meinke PT (2005) PPARs: therapeutic targets for metabolic disease. Trends Pharmacol Sci 26: 244–251. - PubMed
-
- Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, et al. (2001) Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology 142: 4195–4202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

