Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response
- PMID: 23123977
- PMCID: PMC7110875
- DOI: 10.1016/j.micinf.2012.10.008
Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response
Abstract
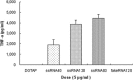
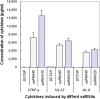
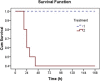
A dangerous cytokine storm occurs in the SARS involving in immune disorder, but many aspects of the pathogenetic mechanism remain obscure since its outbreak. To deeply reveal the interaction of host and SARS-CoV, based on the basic structural feature of pathogen-associated molecular pattern, we created a new bioinformatics method for searching potential pathogenic molecules and identified a set of SARS-CoV specific GU-rich ssRNA fragments with a high-density distribution in the genome. In vitro experiments, the result showed the representative SARS-CoV ssRNAs had powerful immunostimulatory activities to induce considerable level of pro-inflammatory cytokine TNF-a, IL-6 and IL-12 release via the TLR7 and TLR8, almost 2-fold higher than the strong stimulatory ssRNA40 that was found previously from other virus. Moreover, SARS-CoV ssRNA was able to cause acute lung injury in mice with a high mortality rate in vivo experiment. It suggests that SARS-CoV specific GU-rich ssRNA plays a very important role in the cytokine storm associated with a dysregulation of the innate immunity. This study not only presents new evidence about the immunopathologic damage caused by overactive inflammation during the SARS-CoV infection, but also provides a useful clue for a new therapeutic strategy.
Copyright © 2012 Institut Pasteur. Published by Elsevier Masson SAS. All rights reserved.
Figures





References
-
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Li Y., Luo C., Li W., Xu Z., Zeng C., Bi S., Yu J., Wu J., Yang H. Structure-based preliminary analysis of immunity and virulence of SARS coronavirus. Viral Immunol. 2004;17:528–534. - PubMed
-
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. - PubMed
-
- Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K.Y., Ho D.D. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79:2678–2688. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

