Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines
- PMID: 23124062
- PMCID: PMC3489994
- DOI: 10.1101/gad.203786.112
Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines
Abstract
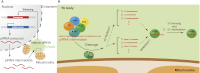
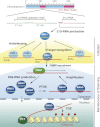
PIWI-interacting RNAs (piRNAs) are endogenous small noncoding RNAs that act as guardians of the genome, protecting it from invasive transposable elements in the germline. Animals lacking piRNA functions show defects in gametogenesis and exhibit sterility. Their descendants are also predisposed to inheriting mutations. Thus, the piRNA pathway has evolved to repress transposons post-transcriptionally and/or transcriptionally. A growing number of studies on piRNAs have investigated piRNA-mediated gene silencing, including piRNA biogenesis. However, piRNAs remain the most enigmatic among all of the silencing-inducing small RNAs because of their complexity and uniqueness. Although piRNAs have been previously suggested to be germline-specific, recent studies have shown that piRNAs also play crucial roles in nongonadal cells. Furthermore, piRNAs have also recently been shown to have roles in multigenerational epigenetic phenomena in worms. The purpose of this review is to highlight new piRNA factors and novel insights in the piRNA world.
Figures


References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 - PubMed
-
- Arkov AL, Wang J-YS, Ramos A, Lehmann R 2006. The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
