cAMP-responsive element modulator α (CREMα) contributes to decreased Notch-1 expression in T cells from patients with active systemic lupus erythematosus (SLE)
- PMID: 23124208
- PMCID: PMC3522254
- DOI: 10.1074/jbc.M112.425371
cAMP-responsive element modulator α (CREMα) contributes to decreased Notch-1 expression in T cells from patients with active systemic lupus erythematosus (SLE)
Abstract
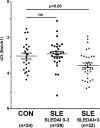
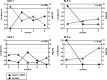
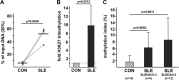
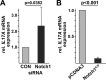
Notch signaling constitutes an evolutionarily conserved pathway that transduces signals between neighboring cells and determines major decisions in cell proliferation, survival, and differentiation. Notch signaling has been shown to play a pivotal role during T cell lineage determination. T lymphocytes from patients with systemic lupus erythematosus (SLE) display a severely altered phenotype with several molecular and functional aberrations, including defective capacities to up-regulate Notch-1 receptor expression upon T cell receptor activation. Here, we demonstrate that basal Notch-1 expression is decreased in T cells from active SLE patients at the mRNA and protein levels in various T cell subpopulations. Notch-1 transcript numbers inversely correlate with disease activity in SLE patients. We provide evidence that both enhanced histone H3 methylation and CpG DNA methylation of the human Notch-1 promoter contribute to decreased Notch-1 expression in SLE T cells. Previous data from our group identified cAMP-responsive element modulator α (CREMα), which is up-regulated in SLE T cells, as a key regulator of epigenetic patterns and gene transcription, e.g. that of IL2 and IL17 genes. In this study, we observed increased CREMα binding to the Notch-1 promoter, which eventually resulted in significantly reduced Notch-1 promoter activity and gene transcription. Notably, decreased Notch-1 levels were associated with elevated IL-17A levels. Our data suggest a role for Notch-1 in SLE immunopathogenesis, and for the first time, we present molecular mechanisms that mediate dysregulated Notch-1 expression in SLE T cells.
Figures


References
-
- Tsokos G. C. (2011) Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 - PubMed
-
- Rauen T., Hedrich C. M., Juang Y. T., Tenbrock K., Tsokos G. C. (2011) cAMP-responsive element modulator (CREM) α protein induces interleukin-17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J. Biol. Chem. 286, 43437–43446 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

