Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat
- PMID: 23124323
- PMCID: PMC3532256
- DOI: 10.1104/pp.112.205161
Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat
Abstract
Cycles of whole-genome duplication (WGD) and diploidization are hallmarks of eukaryotic genome evolution and speciation. Polyploid wheat (Triticum aestivum) has had a massive increase in genome size largely due to recent WGDs. How these processes may impact the dynamics of gene evolution was studied by comparing the patterns of gene structure changes, alternative splicing (AS), and codon substitution rates among wheat and model grass genomes. In orthologous gene sets, significantly more acquired and lost exonic sequences were detected in wheat than in model grasses. In wheat, 35% of these gene structure rearrangements resulted in frame-shift mutations and premature termination codons. An increased codon mutation rate in the wheat lineage compared with Brachypodium distachyon was found for 17% of orthologs. The discovery of premature termination codons in 38% of expressed genes was consistent with ongoing pseudogenization of the wheat genome. The rates of AS within the individual wheat subgenomes (21%-25%) were similar to diploid plants. However, we uncovered a high level of AS pattern divergence between the duplicated homeologous copies of genes. Our results are consistent with the accelerated accumulation of AS isoforms, nonsynonymous mutations, and gene structure rearrangements in the wheat lineage, likely due to genetic redundancy created by WGDs. Whereas these processes mostly contribute to the degeneration of a duplicated genome and its diploidization, they have the potential to facilitate the origin of new functional variations, which, upon selection in the evolutionary lineage, may play an important role in the origin of novel traits.
Figures
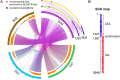

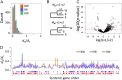
References
-
- Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Deal KR, et al. (2008) Purifying selection and gene conversion in polyploid wheat evolution. In Proceedings of the 11th International Wheat Genetics Symposium. Sydney University Press, Sydney, Australia, pp 1–3
-
- Akhunov ED, Akhunova AR, Dvorak J. (2007) Mechanisms and rates of birth and death of dispersed duplicated genes during the evolution of a multigene family in diploid and tetraploid wheats. Mol Biol Evol 24: 539–550 - PubMed
-
- Akhunov ED, Akhunova AR, Linkiewicz AM, Dubcovsky J, Hummel D, Lazo G, Chao S, Anderson OD, David J, Qi L, et al. (2003) Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc Natl Acad Sci USA 100: 10836–10841 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

