Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3
- PMID: 23124326
- PMCID: PMC3532255
- DOI: 10.1104/pp.112.203166
Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3
Abstract
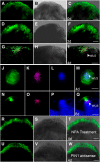
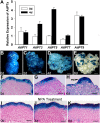
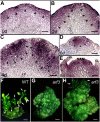
De novo organ regeneration is an excellent biological system for the study of fundamental questions regarding stem cell initiation, cell fate determination, and hormone signaling. Despite the general belief that auxin and cytokinin responses interact to regulate de novo organ regeneration, the molecular mechanisms underlying such a cross talk are little understood. Here, we show that spatiotemporal biosynthesis and polar transport resulted in local auxin distribution in Arabidopsis (Arabidopsis thaliana), which in turn determined the cytokinin response during de novo shoot regeneration. Genetic and pharmacological interference of auxin distribution disrupted the cytokinin response and ATP/ADP ISOPENTENYLTRANSFERASE5 (AtIPT5) expression, affecting stem cell initiation and meristem formation. Transcriptomic data suggested that AUXIN RESPONSE FACTOR3 (ARF3) mediated the auxin response during de novo organ regeneration. Indeed, mutations in ARF3 caused ectopic cytokinin biosynthesis via the misexpression of AtIPT5, and this disrupted organ regeneration. We further showed that ARF3 directly bound to the promoter of AtIPT5 and negatively regulated AtIPT5 expression. The results from this study thus revealed an auxin-cytokinin cross talk mechanism involving distinct intermediate signaling components required for de novo stem cell initiation and shed new light on the mechanisms of organogenesis in planta.
Figures





References
-
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D. (2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J 57: 626–644 - PubMed
-
- Bhojwani SS, Razdan MK. (1996) Plant Tissue Culture: Theory and Practice, a revised edition. Elsevier Press, New York.
-
- Buechel S, Leibfried A, To JP, Zhao Z, Andersen SU, Kieber JJ, Lohmann JU. (2010) Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur J Cell Biol 89: 279–284 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

