Cancer cells preferentially lose small chromosomes
- PMID: 23124507
- PMCID: PMC3587043
- DOI: 10.1002/ijc.27924
Cancer cells preferentially lose small chromosomes
Abstract
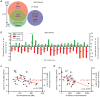
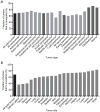
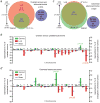
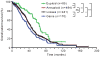
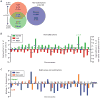
Genetic and genomic aberrations are the primary cause of cancer. Chromosome missegregation leads to aneuploidy and provides cancer cells with a mechanism to lose tumor suppressor loci and gain extra copies of oncogenes. Using cytogenetic and array-based comparative genomic hybridization data, we analyzed numerical chromosome aneuploidy in 43,205 human tumors and found that 68% of solid tumors are aneuploid. In solid tumors, almost all chromosomes are more frequently lost than gained with chromosomes 7, 12 and 20 being the only exceptions with more frequent gains. Strikingly, small chromosomes are lost more readily than large ones, but no such inverse size correlation is observed with chromosome gains. Because of increasing levels of proteotoxic stress, chromosome gains have been shown to slow cell proliferation in a manner proportional to the number of extra gene copies gained. However, we find that the extra chromosome in trisomic tumors does not preferentially have a low gene copy number, suggesting that a proteotoxicity-mediated proliferation barrier is not sustained during tumor progression. Paradoxically, despite a bias toward chromosome loss, gains of chromosomes are a poor prognostic marker in ovarian adenocarcinomas. In addition, we find that solid and non-solid cancers have markedly distinct whole-chromosome aneuploidy signatures, which may underlie their fundamentally different etiologies. Finally, preferential chromosome loss is observed in both early and late stages of astrocytoma. Our results open up new avenues of enquiry into the role and nature of whole-chromosome aneuploidy in human tumors and will redirect modeling and genetic targeting efforts in patients.
Copyright © 2012 UICC.
Figures

References
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–76. - PubMed
-
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. - PubMed
-
- McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

