Global analysis of mRNA stability in Mycobacterium tuberculosis
- PMID: 23125364
- PMCID: PMC3592478
- DOI: 10.1093/nar/gks1019
Global analysis of mRNA stability in Mycobacterium tuberculosis
Abstract
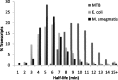
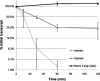
Mycobacterium tuberculosis (MTB) is a highly successful pathogen that infects over a billion people. As with most organisms, MTB adapts to stress by modifying its transcriptional profile. Remodeling of the transcriptome requires both altering the transcription rate and clearing away the existing mRNA through degradation, a process that can be directly regulated in response to stress. To understand better how MTB adapts to the harsh environs of the human host, we performed a global survey of the decay rates of MTB mRNA transcripts. Decay rates were measured for 2139 of the ~4000 MTB genes, which displayed an average half-life of 9.5 min. This is nearly twice the average mRNA half-life of other prokaryotic organisms where these measurements have been made. The transcriptome was further stabilized in response to lowered temperature and hypoxic stress. The generally stable transcriptome described here, and the additional stabilization in response to physiologically relevant stresses, has far-ranging implications for how this pathogen is able to adapt in its human host.
Figures


References
-
- Hambraeus G, von Wachenfeldt C, Hederstedt L. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics. 2003;269:706–714. - PubMed
-
- Cohen SN, McDowall KJ. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 1997;23:1099–1106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

