Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1
- PMID: 23125841
- PMCID: PMC3486646
- DOI: 10.3389/fmicb.2012.00367
Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1
Abstract
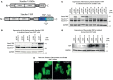
The human immunodeficiency virus type 1 (HIV-1) unspliced, 9 kb genomic RNA (vRNA) is exported from the nucleus for the synthesis of viral structural proteins and enzymes (Gag and Gag/Pol) and is then transported to sites of virus assembly where it is packaged into progeny virions. vRNA co-exists in the cytoplasm in the context of the HIV-1 ribonucleoprotein (RNP) that is currently defined by the presence of Gag and several host proteins including the double-stranded RNA-binding protein, Staufen1. In this study we isolated Staufen1 RNP complexes derived from HIV-1-expressing cells using tandem affinity purification and have identified multiple host protein components by mass spectrometry. Four viral proteins, including Gag, Gag/Pol, Env and Nef as well as >200 host proteins were identified in these RNPs. Moreover, HIV-1 induces both qualitative and quantitative differences in host protein content in these RNPs. 22% of Staufen1-associated factors are virion-associated suggesting that the RNP could be a vehicle to achieve this. In addition, we provide evidence on how HIV-1 modulates the composition of cytoplasmic Staufen1 RNPs. Biochemical fractionation by density gradient analyses revealed new facets on the assembly of Staufen1 RNPs. The assembly of dense Staufen1 RNPs that contain Gag and several host proteins were found to be entirely RNA-dependent but their assembly appeared to be independent of Gag expression. Gag-containing complexes fractionated into a lighter and another, more dense pool. Lastly, Staufen1 depletion studies demonstrated that the previously characterized Staufen1 HIV-1-dependent RNPs are most likely aggregates of smaller RNPs that accumulate at juxtanuclear domains. The molecular characterization of Staufen1 HIV-1 RNPs will offer important information on virus-host cell interactions and on the elucidation of the function of these RNPs for the transport of Gag and the fate of the unspliced vRNA in HIV-1-producing cells.
Keywords: Gag; HIV-1; Staufen1; genomic RNA; gradient centrifugation; mass spectrometry; ribonucleoprotein; virus-host interactions.
Figures






References
LinkOut - more resources
Full Text Sources

