Regulation of human Cripto-1 expression by nuclear receptors and DNA promoter methylation in human embryonal and breast cancer cells
- PMID: 23129342
- PMCID: PMC3573215
- DOI: 10.1002/jcp.24271
Regulation of human Cripto-1 expression by nuclear receptors and DNA promoter methylation in human embryonal and breast cancer cells
Abstract
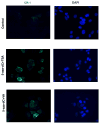
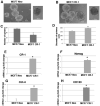
Human Cripto-1 (CR-1) plays an important role in regulating embryonic development while also regulating various stages of tumor progression. However, mechanisms that regulate CR-1 expression during embryogenesis and tumorigenesis are still not well defined. In the present study, we investigated the effects of two nuclear receptors, liver receptor homolog (LRH)-1 and germ cell nuclear factor receptor (GCNF) and epigenetic modifications on CR-1 gene expression in NTERA-2 human embryonal carcinoma cells and in breast cancer cells. CR-1 expression in NTERA-2 cells was positively regulated by LRH-1 through direct binding to a DR0 element within the CR-1 promoter, while GCNF strongly suppressed CR-1 expression in these cells. In addition, the CR-1 promoter was unmethylated in NTERA-2 cells, while T47D, ZR75-1, and MCF7 breast cancer cells showed high levels of CR-1 promoter methylation and low CR-1 mRNA and protein expression. Treatment of breast cancer cells with a demethylating agent and histone deacetylase inhibitors reduced methylation of the CR-1 promoter and reactivated CR-1 mRNA and protein expression in these cells, promoting migration and invasion of breast cancer cells. Analysis of a breast cancer tissue array revealed that CR-1 was highly expressed in the majority of human breast tumors, suggesting that CR-1 expression in breast cancer cell lines might not be representative of in vivo expression. Collectively, these findings offer some insight into the transcriptional regulation of CR-1 gene expression and its critical role in the pathogenesis of human cancer.
Copyright © 2012 Wiley Periodicals, Inc.
Figures









References
-
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, Edel M, Boue S, Izpisua Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26(11):1276–1284. - PubMed
-
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321(5889):699–702. - PubMed
-
- Assou S, Lecarrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot JP, Gasca S, Hovatta O, Hamamah S, Klein B, De Vos J. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. - PMC - PubMed
-
- Bianco C, Adkins HB, Wechselberger C, Seno M, Normanno N, De Luca A, Sun Y, Khan N, Kenney N, Ebert A, Williams KP, Sanicola M, Salomon DS. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial Cells. Mol Cell Biol. 2002;22(8):2586–2597. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

