Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium
- PMID: 23129646
- PMCID: PMC3511728
- DOI: 10.1073/pnas.1212831109
Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium
Erratum in
- Proc Natl Acad Sci U S A. 2013 Jan 20;110(5):1970
Abstract
Chlamydia trachomatis is among the most clinically significant human pathogens, yet their obligate intracellular nature places severe restrictions upon research. Chlamydiae undergo a biphasic developmental cycle characterized by an infectious cell type known as an elementary body (EB) and an intracellular replicative form called a reticulate body (RB). EBs have historically been described as metabolically dormant. A cell-free (axenic) culture system was developed, which showed high levels of metabolic and biosynthetic activity from both EBs and RBs, although the requirements differed for each. EBs preferentially used glucose-6-phosphate as an energy source, whereas RBs required ATP. Both developmental forms showed increased activity when incubated under microaerobic conditions. Incorporation of isotopically labeled amino acids into proteins from both developmental forms indicated unique expression profiles, which were confirmed by genome-wide transcriptional analysis. The described axenic culture system will greatly enhance biochemical and physiological analyses of chlamydiae.
Conflict of interest statement
The authors declare no conflict of interest.
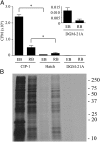
Figures



References
-
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. - PubMed
-
- Unemo M, Clarke IN. The Swedish new variant of Chlamydia trachomatis. Curr Opin Infect Dis. 2011;24:62–69. - PubMed
-
- Shaw EI, et al. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37(4):913–925. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

