FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI
- PMID: 23129770
- PMCID: PMC3527919
- DOI: 10.1074/jbc.M112.414383
FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI
Abstract
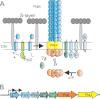
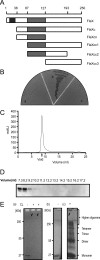
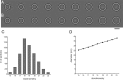
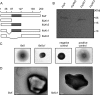
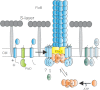
Archaella are the archaeal motility structure, which are structurally similar to gram-negative bacterial type IV pili but functionally resemble bacterial flagella. Structural and biochemical data of archaellum subunits are missing. FlaX, a conserved subunit in crenarchaeal archaella, formed high molecular weight complexes that adapted a ring-like structure with an approximate diameter of 30 nm. The C terminus of FlaX was not only involved in the oligomerization, but also essential for FlaX interaction with FlaI, the bifunctional ATPase that is involved in assembly and rotation of the archaellum. This study gives first insights in the assembly apparatus of archaella.
Figures

References
-
- Jarrell K. F., Albers S. V. (2012) The archaellum. An old motility structure with a new name. Trends Microbiol. 20, 307–312 - PubMed
-
- Ghosh A., Albers S. V. (2011) Assembly and function of the archaeal flagellum. Biochem. Soc. Trans. 39, 64–69 - PubMed
-
- Pohlschroder M., Ghosh A., Tripepi M., Albers S. V. (2011) Archaeal type IV pilus-like structures. Evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363 - PubMed
-
- Lassak K., Neiner T., Ghosh A., Klingl A., Wirth R., Albers S. V. (2012) Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

