Fat body dSir2 regulates muscle mitochondrial physiology and energy homeostasis nonautonomously and mimics the autonomous functions of dSir2 in muscles
- PMID: 23129806
- PMCID: PMC3554107
- DOI: 10.1128/MCB.00976-12
Fat body dSir2 regulates muscle mitochondrial physiology and energy homeostasis nonautonomously and mimics the autonomous functions of dSir2 in muscles
Erratum in
- Mol Cell Biol. 2014 Jul;34(13):2547
Abstract
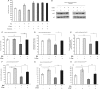
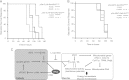
Sir2 is an evolutionarily conserved NAD(+)-dependent deacetylase which has been shown to play a critical role in glucose and fat metabolism. In this study, we have perturbed Drosophila Sir2 (dSir2) expression, bidirectionally, in muscles and the fat body. We report that dSir2 plays a critical role in insulin signaling, glucose homeostasis, and mitochondrial functions. Importantly, we establish the nonautonomous functions of fat body dSir2 in regulating mitochondrial physiology and insulin signaling in muscles. We have identified a novel interplay between dSir2 and dFOXO at an organismal level, which involves Drosophila insulin-like peptide (dILP)-dependent insulin signaling. By genetic perturbations and metabolic rescue, we provide evidence to illustrate that fat body dSir2 mediates its effects on the muscles via free fatty acids (FFA) and dILPs (from the insulin-producing cells [IPCs]). In summary, we show that fat body dSir2 is a master regulator of organismal energy homeostasis and is required for maintaining the metabolic regulatory network across tissues.
Figures



References
-
- Marshall S. 2006. Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci. STKE 2006:re7 doi:10.1126/stke.3462006re7 - DOI - PubMed
-
- Taguchi A, White MF. 2008. Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 70:191–212 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
