ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF
- PMID: 23129809
- PMCID: PMC3554127
- DOI: 10.1128/MCB.01008-12
ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF
Abstract
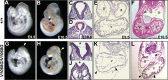
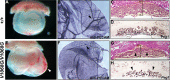
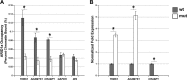
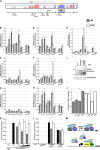
Every known SWI/SNF chromatin-remodeling complex incorporates an ARID DNA binding domain-containing subunit. Despite being a ubiquitous component of the complex, physiological roles for this domain remain undefined. Here, we show that disruption of ARID1a-DNA binding in mice results in embryonic lethality, with mutant embryos manifesting prominent defects in the heart and extraembryonic vasculature. The DNA binding-defective mutant ARID1a subunit is stably expressed and capable of assembling into a SWI/SNF complex with core catalytic properties, but nucleosome substrate binding and promoter occupancy by ARID1a-containing SWI/SNF complexes (BAF-A) are impaired. Depletion of ARID domain-dependent, BAF-A associations at THROMBOSPONDIN 1 (THBS1) led to the concomitant upregulation of this SWI/SNF target gene. Using a THBS1 promoter-reporter gene, we further show that BAF-A directly regulates THBS1 promoter activity in an ARID domain-dependent manner. Our data not only demonstrate that ARID1a-DNA interactions are physiologically relevant in higher eukaryotes but also indicate that these interactions facilitate SWI/SNF binding to target sites in vivo. These findings support the model wherein cooperative interactions among intrinsic subunit-chromatin interaction domains and sequence-specific transcription factors drive SWI/SNF recruitment.
Figures




References
-
- Weintraub H, Groudine M. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193: 848–856 - PubMed
-
- Stern M, Jensen R, Herskowitz I. 1984. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178: 853–868 - PubMed
-
- Cote J, Quinn J, Workman JL, Peterson CL. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
