PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling
- PMID: 23130941
- PMCID: PMC3544699
- DOI: 10.1186/1471-2407-12-507
PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling
Abstract
Background: PCA3 is a non-coding RNA (ncRNA) that is highly expressed in prostate cancer (PCa) cells, but its functional role is unknown. To investigate its putative function in PCa biology, we used gene expression knockdown by small interference RNA, and also analyzed its involvement in androgen receptor (AR) signaling.
Methods: LNCaP and PC3 cells were used as in vitro models for these functional assays, and three different siRNA sequences were specifically designed to target PCA3 exon 4. Transfected cells were analyzed by real-time qRT-PCR and cell growth, viability, and apoptosis assays. Associations between PCA3 and the androgen-receptor (AR) signaling pathway were investigated by treating LNCaP cells with 100 nM dihydrotestosterone (DHT) and with its antagonist (flutamide), and analyzing the expression of some AR-modulated genes (TMPRSS2, NDRG1, GREB1, PSA, AR, FGF8, CdK1, CdK2 and PMEPA1). PCA3 expression levels were investigated in different cell compartments by using differential centrifugation and qRT-PCR.
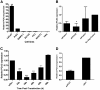
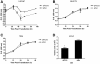
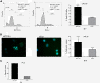
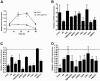
Results: LNCaP siPCA3-transfected cells significantly inhibited cell growth and viability, and increased the proportion of cells in the sub G0/G1 phase of the cell cycle and the percentage of pyknotic nuclei, compared to those transfected with scramble siRNA (siSCr)-transfected cells. DHT-treated LNCaP cells induced a significant upregulation of PCA3 expression, which was reversed by flutamide. In siPCA3/LNCaP-transfected cells, the expression of AR target genes was downregulated compared to siSCr-transfected cells. The siPCA3 transfection also counteracted DHT stimulatory effects on the AR signaling cascade, significantly downregulating expression of the AR target gene. Analysis of PCA3 expression in different cell compartments provided evidence that the main functional roles of PCA3 occur in the nuclei and microsomal cell fractions.
Conclusions: Our findings suggest that the ncRNA PCA3 is involved in the control of PCa cell survival, in part through modulating AR signaling, which may raise new possibilities of using PCA3 knockdown as an additional therapeutic strategy for PCa control.
Figures

References
-
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59(23):5975–5979. - PubMed
-
- de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62(9):2695–2698. - PubMed
-
- Klecka J, Holubec L, Pesta M, Topolcan O, Hora M, Eret V, Finek J, Chottova-Dvorakova M, Babjuk M, Novak K. et al.Differential display code 3 (DD3/PCA3) in prostate cancer diagnosis. Anticancer Res. 2010;30(2):665–670. - PubMed
-
- Neves AF, Araujo TG, Biase WK, Meola J, Alcantara TM, Freitas DG, Goulart LR. Combined analysis of multiple mRNA markers by RT-PCR assay for prostate cancer diagnosis. Clinical Biochem. 2008;41(14–15):1191–1198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

