Lipid monolayer and sparse matrix screening for growing two-dimensional crystals for electron crystallography: methods and examples
- PMID: 23132079
- PMCID: PMC4127334
- DOI: 10.1007/978-1-62703-176-9_28
Lipid monolayer and sparse matrix screening for growing two-dimensional crystals for electron crystallography: methods and examples
Abstract
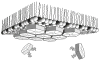
Electron microscopy provides an efficient method for rapidly assessing whether a solution of macromolecules is homogeneous and monodisperse. If the macromolecules can be induced to form two-dimensional crystals that are a single layer in thickness, then electron crystallography of frozen-hydrated crystals has the potential of achieving three-dimensional density maps at sub-nanometer or even atomic resolution. Here we describe the lipid monolayer and sparse matrix screening methods for growing two-dimensional crystals and present successful applications to soluble macromolecular complexes: carboxysome shell proteins and HIV CA, respectively. Since it is common to express recombinant proteins with poly-His tags for purification by metal affinity chromatography, the monolayer technique using bulk lipids doped with Ni(2+) lipids has the potential for broad application. Likewise, the sparse matrix method uses screening conditions for three-dimensional crystallization and is therefore of broad applicability.
Figures






References
-
- McPherson A. Preparation and analysis of protein crystals. Wiley; New York: 1982.
-
- Yeager M, Unger VM, Mitra AK. Three-dimensional structure of membrane proteins determined by two-dimensional crystallization, electron cryomicroscopy, and image analysis. Methods Enzymol. 1999;294:135–180. - PubMed
-
- Adair BD, Yeager M. Electron microscopy of integrins. Methods Enzymol. 2007;426:337–373. - PubMed
-
- Hemming SA, Bochkarev A, Darst SA, Kornberg RD, Ala P, Yang DSC, Edwards AM. The mechanism of protein crystal growth from lipid layers. J Mol Biol. 1995;246:308–316. - PubMed
-
- Brisson A, Olofsson A, Ringler P, Schmutz M, Stoylova S. Two-dimensional crystallization of proteins on planar lipid films and structure determination by electron crystallography. Biol Cell. 1994;80:221–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

