A travel guide to Cytoscape plugins
- PMID: 23132118
- PMCID: PMC3649846
- DOI: 10.1038/nmeth.2212
A travel guide to Cytoscape plugins
Abstract
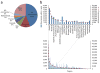
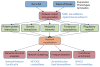
Cytoscape is open-source software for integration, visualization and analysis of biological networks. It can be extended through Cytoscape plugins, enabling a broad community of scientists to contribute useful features. This growth has occurred organically through the independent efforts of diverse authors, yielding a powerful but heterogeneous set of tools. We present a travel guide to the world of plugins, covering the 152 publicly available plugins for Cytoscape 2.5-2.8. We also describe ongoing efforts to distribute, organize and maintain the quality of the collection.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Yeung N, Cline MS, Kuchinsky A, Smoot ME, Bader GD. Exploring biological networks with Cytoscape software. Curr Protoc Bioinformatics. 2008;23:8.13. - PubMed
-
- Hermjakob H, et al. The HUPO PSI’s molecular interaction format—a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22:177–183. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

