Universal pattern and diverse strengths of successive synonymous codon bias in three domains of life, particularly among prokaryotic genomes
- PMID: 23132389
- PMCID: PMC3514858
- DOI: 10.1093/dnares/dss027
Universal pattern and diverse strengths of successive synonymous codon bias in three domains of life, particularly among prokaryotic genomes
Abstract
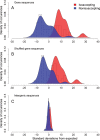
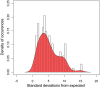
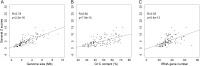
There has been significant progress in understanding the process of protein translation in recent years. One of the best examples is the discovery of usage bias in successive synonymous codons and its role in eukaryotic translation efficiency. We observed here a similar type of bias in the other two life domains, bacteria and archaea, although the bias strength was much smaller than in eukaryotes. Among 136 prokaryotic genomes, 98 were found to have significant bias from random use of successive synonymous codons with Z scores larger than three. Furthermore, significantly different bias strengths were found between prokaryotes grouped by various genomic or biochemical characteristics. Interestingly, the bias strength measured by a general Z score could be fitted well (R = 0.83, P < 10(-15)) by three genomic variables: genome size, G + C content, and tRNA gene number based on multiple linear regression. A different distribution of synonymous codon pairs between protein-coding genes and intergenic sequences suggests that bias is caused by translation selection. The present results indicate that protein translation is tuned by codon (pair) usage, and the intensity of the regulation is associated with genome size, tRNA gene number, and G + C content.
Figures
Similar articles
-
Codon Usage Optimization in the Prokaryotic Tree of Life: How Synonymous Codons Are Differentially Selected in Sequence Domains with Different Expression Levels and Degrees of Conservation.mBio. 2020 Jul 21;11(4):e00766-20. doi: 10.1128/mBio.00766-20. mBio. 2020. PMID: 32694138 Free PMC article.
-
Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency.PLoS One. 2012;7(3):e33547. doi: 10.1371/journal.pone.0033547. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432034 Free PMC article.
-
Codon usage bias in prokaryotic pyrimidine-ending codons is associated with the degeneracy of the encoded amino acids.Nucleic Acids Res. 2012 Aug;40(15):7074-83. doi: 10.1093/nar/gks348. Epub 2012 May 11. Nucleic Acids Res. 2012. PMID: 22581775 Free PMC article.
-
Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes.FEBS Lett. 2010 Jan 21;584(2):252-64. doi: 10.1016/j.febslet.2009.11.052. FEBS Lett. 2010. PMID: 19931533 Review.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
Cited by
-
Mutational Pressure in Zika Virus: Local ADAR-Editing Areas Associated with Pauses in Translation and Replication.Front Cell Infect Microbiol. 2017 Feb 22;7:44. doi: 10.3389/fcimb.2017.00044. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28275585 Free PMC article.
-
Codon selection reduces GC content bias in nucleic acids encoding for intrinsically disordered proteins.Cell Mol Life Sci. 2020 Jan;77(1):149-160. doi: 10.1007/s00018-019-03166-6. Epub 2019 Jun 7. Cell Mol Life Sci. 2020. PMID: 31175370 Free PMC article.
-
Differential bicodon usage in lowly and highly abundant proteins.PeerJ. 2017 Mar 9;5:e3081. doi: 10.7717/peerj.3081. eCollection 2017. PeerJ. 2017. PMID: 28289571 Free PMC article.
-
Dicodon-based measures for modeling gene expression.Bioinformatics. 2023 Jun 1;39(6):btad380. doi: 10.1093/bioinformatics/btad380. Bioinformatics. 2023. PMID: 37307098 Free PMC article.
-
Comprehensive profiling of codon usage signatures and codon context variations in the genus Ustilago.World J Microbiol Biotechnol. 2019 Jul 22;35(8):118. doi: 10.1007/s11274-019-2693-y. World J Microbiol Biotechnol. 2019. PMID: 31332540
References
-
- Cannarozzi G., Schraudolph N.N., Faty M., et al. A role for codon order in translation dynamics. Cell. 2010;141:355–67. doi:10.1016/j.cell.2010.02.036. - DOI - PubMed
-
- Fredrick K., Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–9. doi:10.1016/j.cell.2010.03.033. - DOI - PMC - PubMed
-
- Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–8. doi:10.1126/science.1170160. - DOI - PMC - PubMed
-
- Plotkin J.B., Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi:10.1038/nrg2899. - DOI - PMC - PubMed
-
- Tuller T., Carmi A., Vestsigian K., et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–54. doi:10.1016/j.cell.2010.03.031. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

