The effect of bacterial recombination on adaptation on fitness landscapes with limited peak accessibility
- PMID: 23133344
- PMCID: PMC3487459
- DOI: 10.1371/journal.pcbi.1002735
The effect of bacterial recombination on adaptation on fitness landscapes with limited peak accessibility
Abstract
There is ample empirical evidence revealing that fitness landscapes are often complex: the fitness effect of a newly arisen mutation can depend strongly on the allelic state at other loci. However, little is known about the effects of recombination on adaptation on such fitness landscapes. Here, we investigate how recombination influences the rate of adaptation on a special type of complex fitness landscapes. On these landscapes, the mutational trajectories from the least to the most fit genotype are interrupted by genotypes with low relative fitness. We study the dynamics of adapting populations on landscapes with different compositions and numbers of low fitness genotypes, with and without recombination. Our results of the deterministic model (assuming an infinite population size) show that recombination generally decelerates adaptation on these landscapes. However, in finite populations, this deceleration is outweighed by the accelerating Fisher-Muller effect under certain conditions. We conclude that recombination has complex effects on adaptation that are highly dependent on the particular fitness landscape, population size and recombination rate.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
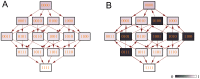
 .
.
 ), B) positive baseline epistasis (
), B) positive baseline epistasis ( ), C) negative baseline epistasis (
), C) negative baseline epistasis ( ). Each box shows the distribution of Tfix across all fitness landscapes with the respective number of LFGs. The boxes give the interquartile range. Outliers are represented with the points in more than 1.5 times the interquartile range from the end of the boxes. The whiskers are extended to the farthest points from the end of the boxes that are not outliers. The black line connects the median of the boxes. The red dashed lines show Tfix on the landscape with no LFG with the corresponding baseline epistasis. In the absence of baseline epistasis and LFGs in the fitness landscape, recombination has no effect on the rate of adaptation (Tfix
). Each box shows the distribution of Tfix across all fitness landscapes with the respective number of LFGs. The boxes give the interquartile range. Outliers are represented with the points in more than 1.5 times the interquartile range from the end of the boxes. The whiskers are extended to the farthest points from the end of the boxes that are not outliers. The black line connects the median of the boxes. The red dashed lines show Tfix on the landscape with no LFG with the corresponding baseline epistasis. In the absence of baseline epistasis and LFGs in the fitness landscape, recombination has no effect on the rate of adaptation (Tfix
 , orange dashed lines). Parameters take the values
, orange dashed lines). Parameters take the values  .
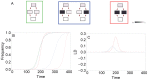
.
 values of 0.95, 1.0 and 1.05, respectively. C) Effect of mutation rate. Red, orange and green curves correspond to
values of 0.95, 1.0 and 1.05, respectively. C) Effect of mutation rate. Red, orange and green curves correspond to  values of 10−6, 10−5 and 10−4, respectively. D) Effect of selection coefficient. Orange, red and green curves correspond to
values of 10−6, 10−5 and 10−4, respectively. D) Effect of selection coefficient. Orange, red and green curves correspond to  values of 0.050, 0.075 and 0.1, respectively. Note the different scales of the y-axes in plots A to D.
values of 0.050, 0.075 and 0.1, respectively. Note the different scales of the y-axes in plots A to D.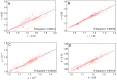
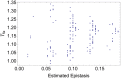
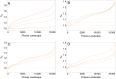
 . See main text for a description of how we measured physiological epistasis on these fitness landscapes.
. See main text for a description of how we measured physiological epistasis on these fitness landscapes.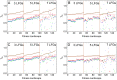
 (red),
(red),  (blue) and
(blue) and  (green). All Tfix values are sorted according to their recombination effect in the deterministic model (brown). Parameters take standard values (see also Figures 4 and 5), and in plots B to D we varies one of the parameters: A) Standard parameter set, B)
(green). All Tfix values are sorted according to their recombination effect in the deterministic model (brown). Parameters take standard values (see also Figures 4 and 5), and in plots B to D we varies one of the parameters: A) Standard parameter set, B)  , C)
, C)  and D)
and D)  .
.Similar articles
-
Recombination accelerates adaptation on a large-scale empirical fitness landscape in HIV-1.PLoS Genet. 2014 Jun 26;10(6):e1004439. doi: 10.1371/journal.pgen.1004439. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24967626 Free PMC article.
-
Recombination and mutational robustness in neutral fitness landscapes.PLoS Comput Biol. 2019 Aug 15;15(8):e1006884. doi: 10.1371/journal.pcbi.1006884. eCollection 2019 Aug. PLoS Comput Biol. 2019. PMID: 31415555 Free PMC article.
-
Evolutionary dynamics of the most populated genotype on rugged fitness landscapes.Phys Rev E Stat Nonlin Soft Matter Phys. 2007 Sep;76(3 Pt 1):031922. doi: 10.1103/PhysRevE.76.031922. Epub 2007 Sep 26. Phys Rev E Stat Nonlin Soft Matter Phys. 2007. PMID: 17930286
-
Empirical fitness landscapes and the predictability of evolution.Nat Rev Genet. 2014 Jul;15(7):480-90. doi: 10.1038/nrg3744. Epub 2014 Jun 10. Nat Rev Genet. 2014. PMID: 24913663 Review.
-
How closely does genetic diversity in finite populations conform to predictions of neutral theory? Large deficits in regions of low recombination.Heredity (Edinb). 2012 Mar;108(3):167-78. doi: 10.1038/hdy.2011.66. Epub 2011 Aug 31. Heredity (Edinb). 2012. PMID: 21878983 Free PMC article. Review.
Cited by
-
Extensive genomic variation within clonal bacterial groups resulted from homologous recombination.Mob Genet Elements. 2013 Jan 1;3(1):e23463. doi: 10.4161/mge.23463. Mob Genet Elements. 2013. PMID: 23734294 Free PMC article.
-
On the deformability of an empirical fitness landscape by microbial evolution.Proc Natl Acad Sci U S A. 2018 Oct 30;115(44):11286-11291. doi: 10.1073/pnas.1808485115. Epub 2018 Oct 15. Proc Natl Acad Sci U S A. 2018. PMID: 30322921 Free PMC article.
-
Recombination accelerates adaptation on a large-scale empirical fitness landscape in HIV-1.PLoS Genet. 2014 Jun 26;10(6):e1004439. doi: 10.1371/journal.pgen.1004439. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24967626 Free PMC article.
-
Evolution at 'Sutures' and 'Centers': Recombination Can Aid Adaptation of Spatially Structured Populations on Rugged Fitness Landscapes.PLoS Comput Biol. 2016 Dec 14;12(12):e1005247. doi: 10.1371/journal.pcbi.1005247. eCollection 2016 Dec. PLoS Comput Biol. 2016. PMID: 27973606 Free PMC article.
-
Emergence and widespread circulation of a recombinant SARS-CoV-2 lineage in North America.Cell Host Microbe. 2022 Aug 10;30(8):1112-1123.e3. doi: 10.1016/j.chom.2022.06.010. Epub 2022 Jun 20. Cell Host Microbe. 2022. PMID: 35853454 Free PMC article.
References
-
- Otto SP, Lenormand T (2002) Resolving the paradox of sex and recombination. Nature Rev Genet 3: 252–261. - PubMed
-
- Michod RE, Levin BR (1987) The Evolution of Sex: An Examination of Current Ideas. Michigan: Sinauer Associates. 352 p.
-
- Weinreich DM, Watson RA, Chao L (2005) Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59: 1165–1174. - PubMed
-
- Eshel I, Feldman MW (1970) On the evolutionary effect of recombination. Theor Popul Biol 1: 88–100. - PubMed
-
- Kondrashov AS (1988) Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

